Multiomics integration-based molecular characterizations of COVID-19
- PMID: 34864875
- PMCID: PMC8769889
- DOI: 10.1093/bib/bbab485
Multiomics integration-based molecular characterizations of COVID-19
Abstract
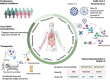
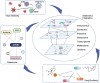
The coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), rapidly became a global health challenge, leading to unprecedented social and economic consequences. The mechanisms behind the pathogenesis of SARS-CoV-2 are both unique and complex. Omics-scale studies are emerging rapidly and offer a tremendous potential to unravel the puzzle of SARS-CoV-2 pathobiology, as well as moving forward with diagnostics, potential drug targets, risk stratification, therapeutic responses, vaccine development and therapeutic innovation. This review summarizes various aspects of understanding multiomics integration-based molecular characterizations of COVID-19, which to date include the integration of transcriptomics, proteomics, genomics, lipidomics, immunomics and metabolomics to explore virus targets and developing suitable therapeutic solutions through systems biology tools. Furthermore, this review also covers an abridgment of omics investigations related to disease pathogenesis and virulence, the role of host genetic variation and a broad array of immune and inflammatory phenotypes contributing to understanding COVID-19 traits. Insights into this review, which combines existing strategies and multiomics integration profiling, may help further advance our knowledge of COVID-19.
Keywords: COVID-19; molecular characteristics; multiomics integration; outcome; severity; single-cell omics.
© The Author(s) 2021. Published by Oxford University Press.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

