Genomic signatures and insights into host niche adaptation of the entomopathogenic fungus Metarhizium humberi
- PMID: 34865006
- PMCID: PMC9210286
- DOI: 10.1093/g3journal/jkab416
Genomic signatures and insights into host niche adaptation of the entomopathogenic fungus Metarhizium humberi
Abstract
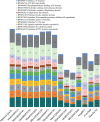
The genus Metarhizium is composed of species used in biological control programs of agricultural pests worldwide. This genus includes common fungal pathogen of many insects and mites and endophytes that can increase plant growth. Metarhizium humberi was recently described as a new species. This species is highly virulent against some insect pests and promotes growth in sugarcane, strawberry, and soybean crops. In this study, we sequenced the genome of M. humberi, isolate ESALQ1638, and performed a functional analysis to determine its genomic signatures and highlight the genes and biological processes associated with its lifestyle. The genome annotation predicted 10633 genes in M. humberi, of which 92.0% are assigned putative functions, and ∼17% of the genome was annotated as repetitive sequences. We found that 18.5% of the M. humberi genome is similar to experimentally validated proteins associated with pathogen-host interaction. Compared to the genomes of eight Metarhizium species, the M. humberi ESALQ1638 genome revealed some unique traits that stood out, e.g., more genes functionally annotated as polyketide synthases (PKSs), overrepresended GO-terms associated to transport of ions, organic and amino acid, a higher percentage of repetitive elements, and higher levels of RIP-induced point mutations. The M. humberi genome will serve as a resource for promoting studies on genome structure and evolution that can contribute to research on biological control and plant biostimulation. Thus, the genomic data supported the broad host range of this species within the generalist PARB clade and suggested that M. humberi ESALQ1638 might be particularly good at producing secondary metabolites and might be more efficient in transporting amino acids and organic compounds.
Keywords: entomopathogenic fungus; genomic features; host adaptation; specialization.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures




References
-
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J.. 2004. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 20:578–580. - PubMed
-
- Andrews S. 2010. Babraham bioinformatics-FastQC a quality control tool for high throughput sequence data. URL: https://www. bioinformatics. babraham. ac. uk/projects/fastqc.
-
- Bagga S, Hu G, Screen SE, St Leger RJ.. 2004. Reconstructing the diversification of subtilisins in the pathogenic fungus Metarhizium anisopliae. Gene. 324:159–169. - PubMed
-
- Barelli L, Moonjely S, Behie SW, Bidochka MJ.. 2016. Fungi with multifunctional lifestyles: endophytic insect pathogenic fungi. Plant Mol Biol. 90:657–664. - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous
