The auxin-inducible degron 2 (AID2) system enables controlled protein knockdown during embryogenesis and development in Caenorhabditis elegans
- PMID: 34865044
- PMCID: PMC9208642
- DOI: 10.1093/genetics/iyab218
The auxin-inducible degron 2 (AID2) system enables controlled protein knockdown during embryogenesis and development in Caenorhabditis elegans
Abstract
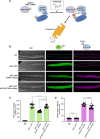
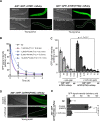
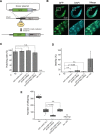
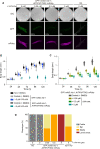
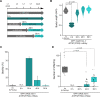
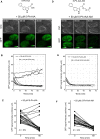
Targeted protein degradation using the auxin-inducible degron (AID) system is garnering attention in the research field of Caenorhabditis elegans, because of the rapid and efficient target depletion it affords, which can be controlled by treating the animals with the phytohormone auxin. However, the current AID system has drawbacks, i.e., leaky degradation in the absence of auxin and the requirement for high auxin doses. Furthermore, it is challenging to deplete degron-fused proteins in embryos because of their eggshell, which blocks auxin permeability. Here, we apply an improved AID2 system utilizing AtTIR1(F79G) and 5-phenyl-indole-3-acetic acid (5-Ph-IAA) to C. elegans and demonstrated that it confers better degradation control vs the previous system by suppressing leaky degradation and inducing sharp degradation using 1,300-fold lower 5-Ph-IAA doses. We successfully degraded the endogenous histone H2A.Z protein fused to an mAID degron and disclosed its requirement in larval growth and reproduction, regardless of the presence of maternally inherited H2A.Z molecules. Moreover, we developed an eggshell-permeable 5-Ph-IAA analog, 5-Ph-IAA-AM, that affords an enhanced degradation in laid embryos. Our improved system will contribute to the disclosure of the roles of proteins in C. elegans, in particular those that are involved in embryogenesis and development, through temporally controlled protein degradation.
Keywords: Caenorhabditis elegans; 5-Ph-IAA; 5-Ph-IAA-AM; AID; auxin; degron; histone H2A.Z; protein knockdown; targeted protein degradation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

