Rice Cell Division Cycle 20s are required for faithful chromosome segregation and cytokinesis during meiosis
- PMID: 34865119
- PMCID: PMC8825277
- DOI: 10.1093/plphys/kiab543
Rice Cell Division Cycle 20s are required for faithful chromosome segregation and cytokinesis during meiosis
Abstract
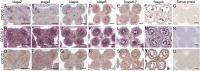
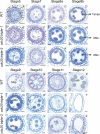
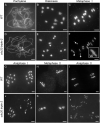
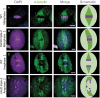
Chromosome segregation must be under strict regulation to maintain chromosome euploidy and stability. Cell Division Cycle 20 (CDC20) is an essential cell cycle regulator that promotes the metaphase-to-anaphase transition and functions in the spindle assembly checkpoint, a surveillance pathway that ensures the fidelity of chromosome segregation. Plant CDC20 genes are present in multiple copies, and whether CDC20s have the same functions in plants as in yeast and animals is unclear, given the potential for divergence or redundancy among the multiple copies. Here, we studied all three CDC20 genes in rice (Oryza sativa) and constructed two triple mutants by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9-mediated genome editing to explore their roles in development. Knocking out all three CDC20 genes led to total sterility but did not affect vegetative development. Loss of the three CDC20 proteins did not alter mitotic division but severely disrupted meiosis as a result of asynchronous and unequal chromosome segregation, chromosome lagging, and premature separation of chromatids. Immunofluorescence of tubulin revealed malformed meiotic spindles in microsporocytes of the triple mutants. Furthermore, cytokinesis of meiosis I was absent or abnormal, and cytokinesis II was completely prevented in all mutant microsporocytes; thus, no tetrads or pollen formed in either cdc20 triple mutant. Finally, the subcellular structures and functions of the tapetum were disturbed by the lack of CDC20 proteins. These findings demonstrate that the three rice CDC20s play redundant roles but are indispensable for faithful meiotic chromosome segregation and cytokinesis, which are required for the production of fertile microspores.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




Similar articles
-
ANAPHASE-PROMOTING COMPLEX/CYCLOSOME coactivators maintain AURORA 1 kinase homeostasis during meiotic chromosome segregation.Plant Cell. 2025 Jun 4;37(6):koaf089. doi: 10.1093/plcell/koaf089. Plant Cell. 2025. PMID: 40244922 Free PMC article.
-
Arabidopsis Cell Division Cycle 20.1 Is Required for Normal Meiotic Spindle Assembly and Chromosome Segregation.Plant Cell. 2015 Dec;27(12):3367-82. doi: 10.1105/tpc.15.00834. Epub 2015 Dec 15. Plant Cell. 2015. PMID: 26672070 Free PMC article.
-
Heat stress interferes with chromosome segregation and cytokinesis during male meiosis in Arabidopsis thaliana.Plant Signal Behav. 2020 May 3;15(5):1746985. doi: 10.1080/15592324.2020.1746985. Epub 2020 Apr 10. Plant Signal Behav. 2020. PMID: 32275182 Free PMC article.
-
Spindle formation, chromosome segregation and the spindle checkpoint in mammalian oocytes and susceptibility to meiotic error.Mutat Res. 2008 Mar 12;651(1-2):14-29. doi: 10.1016/j.mrgentox.2007.10.015. Epub 2007 Nov 9. Mutat Res. 2008. PMID: 18096427 Review.
-
Meiotic prophase-like pathway for cleavage-independent removal of cohesin for chromosome morphogenesis.Curr Genet. 2019 Aug;65(4):817-827. doi: 10.1007/s00294-019-00959-x. Epub 2019 Mar 28. Curr Genet. 2019. PMID: 30923890 Review.
Cited by
-
Biochemical, biophysical, and functional characterisation of the E3 ubiquitin ligase APC/C regulator CDC20 from Arabidopsis thaliana.Front Physiol. 2022 Jul 25;13:938688. doi: 10.3389/fphys.2022.938688. eCollection 2022. Front Physiol. 2022. PMID: 35957989 Free PMC article.
-
ANAPHASE-PROMOTING COMPLEX/CYCLOSOME coactivators maintain AURORA 1 kinase homeostasis during meiotic chromosome segregation.Plant Cell. 2025 Jun 4;37(6):koaf089. doi: 10.1093/plcell/koaf089. Plant Cell. 2025. PMID: 40244922 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

