Nucleosomes enter cells by clathrin- and caveolin-dependent endocytosis
- PMID: 34865123
- PMCID: PMC8643636
- DOI: 10.1093/nar/gkab1121
Nucleosomes enter cells by clathrin- and caveolin-dependent endocytosis
Abstract
DNA damage and apoptosis lead to the release of free nucleosomes-the basic structural repeating units of chromatin-into the blood circulation system. We recently reported that free nucleosomes that enter the cytoplasm of mammalian cells trigger immune responses by activating cGMP-AMP synthase (cGAS). In the present study, we designed experiments to reveal the mechanism of nucleosome uptake by human cells. We showed that nucleosomes are first absorbed on the cell membrane through nonspecific electrostatic interactions between positively charged histone N-terminal tails and ligands on the cell surface, followed by internalization via clathrin- or caveolae-dependent endocytosis. After cellular internalization, endosomal escape occurs rapidly, and nucleosomes are released into the cytosol, maintaining structural integrity for an extended period. The efficient endocytosis of extracellular nucleosomes suggests that circulating nucleosomes may lead to cellular disorders as well as immunostimulation, and thus, the biological effects exerted by endocytic nucleosomes should be addressed in the future.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

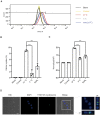
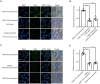
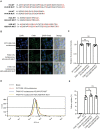
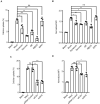
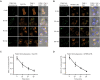
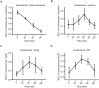
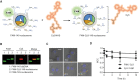
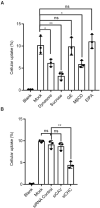
Similar articles
-
A distinct class of endosome mediates clathrin-independent endocytosis to the Golgi complex.Nat Cell Biol. 2002 May;4(5):374-8. doi: 10.1038/ncb787. Nat Cell Biol. 2002. PMID: 11951093
-
Glycosphingolipids internalized via caveolar-related endocytosis rapidly merge with the clathrin pathway in early endosomes and form microdomains for recycling.J Biol Chem. 2003 Feb 28;278(9):7564-72. doi: 10.1074/jbc.M210457200. Epub 2002 Dec 12. J Biol Chem. 2003. PMID: 12482757
-
Distinct caveolae-mediated endocytic pathways target the Golgi apparatus and the endoplasmic reticulum.J Cell Sci. 2003 Mar 15;116(Pt 6):1059-71. doi: 10.1242/jcs.00327. J Cell Sci. 2003. PMID: 12584249
-
Transport of protein toxins into cells: pathways used by ricin, cholera toxin and Shiga toxin.FEBS Lett. 2002 Oct 2;529(1):49-53. doi: 10.1016/s0014-5793(02)03182-4. FEBS Lett. 2002. PMID: 12354612 Review.
-
Sorting signals.Curr Opin Cell Biol. 1989 Aug;1(4):617-23. doi: 10.1016/0955-0674(89)90024-0. Curr Opin Cell Biol. 1989. PMID: 2576381 Free PMC article. Review. No abstract available.
Cited by
-
Extracellular Histones Activate Endothelial NLRP3 Inflammasome and are Associated with a Severe Sepsis Phenotype.J Inflamm Res. 2022 Jul 25;15:4217-4238. doi: 10.2147/JIR.S363693. eCollection 2022. J Inflamm Res. 2022. PMID: 35915852 Free PMC article.
-
LY6K depletion modulates TGF-β and EGF signaling.Cancer Med. 2023 Jun;12(11):12593-12607. doi: 10.1002/cam4.5940. Epub 2023 Apr 19. Cancer Med. 2023. PMID: 37076981 Free PMC article.
-
Circulating Nucleosomes as a Novel Biomarker for Sepsis: A Scoping Review.Biomedicines. 2024 Jun 21;12(7):1385. doi: 10.3390/biomedicines12071385. Biomedicines. 2024. PMID: 39061959 Free PMC article.
-
An integrative analysis reveals cancer risk associated with artificial sweeteners.J Transl Med. 2025 Jan 8;23(1):32. doi: 10.1186/s12967-024-06047-0. J Transl Med. 2025. PMID: 39780215 Free PMC article.
-
Pharmacological effects, molecular mechanisms and strategies to improve bioavailability of curcumin in the treatment of neurodegenerative diseases.Front Pharmacol. 2025 Jul 10;16:1625821. doi: 10.3389/fphar.2025.1625821. eCollection 2025. Front Pharmacol. 2025. PMID: 40709087 Free PMC article. Review.
References
-
- Holdenrieder S., Stieber P., Bodenmuller H., Busch M., Pawel J., Schalhorn A., Nagel D., Seidel D.. Circulating nucleosomes in serum. Ann. N. Y. Acad. Sci. 2001; 945:93–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

