A Lipidomic Approach to Identify Potential Biomarkers in Exosomes From Melanoma Cells With Different Metastatic Potential
- PMID: 34867454
- PMCID: PMC8637280
- DOI: 10.3389/fphys.2021.748895
A Lipidomic Approach to Identify Potential Biomarkers in Exosomes From Melanoma Cells With Different Metastatic Potential
Abstract
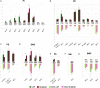
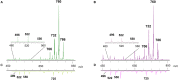
Melanoma, one of the most lethal cutaneous cancers, is characterized by its ability to metastasize to other distant sites, such as the bone. Melanoma cells revealed a variable in vitro propensity to be attracted toward bone fragments, and melanoma-derived exosomes play a role in regulating the osteotropism of these cells. We have here investigated the lipid profiles of melanoma cell lines (LCP and SK-Mel28) characterized by different metastatic propensities to colonize the bone. We have purified exosomes from cell supernatants by ultracentrifugation, and their lipid composition has been compared to identify potential lipid biomarkers for different migration and invasiveness of melanoma cells. Matrix-assisted laser desorption ionization-time-of-flight/mass spectrometry (MALDI-TOF/MS) lipid analysis has been performed on very small amounts of intact parental cells and exosomes by skipping lipid extraction and separation steps. Statistical analysis has been applied to MALDI mass spectra in order to discover significant differences in lipid profiles. Our results clearly show more saturated and shorter fatty acid tails in poorly metastatic (LCP) cells compared with highly metastatic (SK-Mel28) cells, particularly for some species of phosphatidylinositol. Sphingomyelin, lysophosphatidylcholine, and phosphatidic acid were enriched in exosome membranes compared to parental cells. In addition, we have clearly detected a peculiar phospholipid bis(monoacylglycero)phosphate as a specific lipid marker of exosomes. MALDI-TOF/MS lipid profiles of exosomes derived from the poorly and highly metastatic cells were not significantly different.
Keywords: MALDI-TOF/MS; lipids; melanoma; membrane vesicles; osteotropism.
Copyright © 2021 Lobasso, Tanzarella, Mannavola, Tucci, Silvestris, Felici, Ingrosso, Corcelli and Lopalco.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Anderson D. M. G., Ablonczy Z., Koutalos Y., Hanneken A. M., Spraggins J. M., Calcutt M. W., et al. (2017). Bis(monoacylglycero)phosphate lipids in the retinal pigment epithelium implicate lysosomal/endosomal dysfunction in a model of Stargardt disease and human retinas. Sci. Rep. 7:17352. 10.1038/s41598-017-17402-1 - DOI - PMC - PubMed
-
- Bligh E. G., Dyer W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37 911–917. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

