Optimization and Clinical Validation of Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification, a Fast, Highly Sensitive and Specific COVID-19 Molecular Diagnostic Tool That Is Robust to Detect SARS-CoV-2 Variants of Concern
- PMID: 34867841
- PMCID: PMC8637279
- DOI: 10.3389/fmicb.2021.713713
Optimization and Clinical Validation of Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification, a Fast, Highly Sensitive and Specific COVID-19 Molecular Diagnostic Tool That Is Robust to Detect SARS-CoV-2 Variants of Concern
Abstract
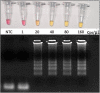
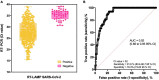
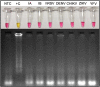
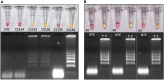
The coronavirus disease 2019 (COVID-19) pandemic unfolded due to the widespread severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission reinforced the urgent need for affordable molecular diagnostic alternative methods for massive testing screening. We present the clinical validation of a pH-dependent colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) for SARS-CoV-2 detection. The method revealed a limit of detection of 19.3 ± 2.7 viral genomic copies/μL when using RNA extracted samples obtained from nasopharyngeal swabs collected in guanidine-containing viral transport medium. Typical RT-LAMP reactions were performed at 65°C for 30 min. When compared to reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR), up to cycle-threshold (Ct) value 32, RT-LAMP presented 98% [95% confidence interval (CI) = 95.3-99.5%] sensitivity and 100% (95% CI = 94.5-100%) specificity for SARS-CoV-2 RNA detection targeting E and N genes. No cross-reactivity was detected when testing other non-SARS-CoV virus, confirming high specificity. The test is compatible with primary RNA extraction-free samples. We also demonstrated that colorimetric RT-LAMP can detect SARS-CoV-2 variants of concern and variants of interest, such as variants occurring in Brazil named gamma (P.1), zeta (P.2), delta (B.1.617.2), B.1.1.374, and B.1.1.371. The method meets point-of-care requirements and can be deployed in the field for high-throughput COVID-19 testing campaigns, especially in countries where COVID-19 testing efforts are far from ideal to tackle the pandemics. Although RT-qPCR is considered the gold standard for SARS-CoV-2 RNA detection, it requires expensive equipment, infrastructure, and highly trained personnel. In contrast, RT-LAMP emerges as an affordable, inexpensive, and simple alternative for SARS-CoV-2 molecular detection that can be applied to massive COVID-19 testing campaigns and save lives.
Keywords: COVID-19; RT-LAMP; SARS-CoV-2; diagnostic test; molecular test; respiratory virus.
Copyright © 2021 Alves, de Oliveira, Franco-Luiz, Almeida, Gonçalves, Borges, Rocha, Rocha, Bezerra, Miranda, Capanema, Martins, Weber, Teixeira, Wallau and do Monte-Neto.
Conflict of interest statement
HM is part of Visuri company. Results presented here are the basis of a COVID-19 RT-LAMP diagnostic test offered by Visuri named OmniLAMP® SARS-CoV-2 kit. PA and RM-N are co-founders and scientific advisors at CEPHA Biotech. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Anahtar M. N., McGrath G. E. G., Rabe B. A., Tanner N. A., White B. A., Lennerz J. K. M., et al. (2021). Clinical assessment and validation of a rapid and sensitive SARS-CoV-2 test using reverse transcription loop-mediated isothermal amplification without the need for RNA extraction. Open Forum Infect. Dis. 8:ofaa631. 10.1093/ofid/ofaa631 - DOI - PMC - PubMed
-
- Asprino P., Bettoni F., Camargo A., Coelho D., Coppini G., Correa I., et al. (2020). A scalable saliva-based, extraction-free rt-lamp protocol for sars-cov-2 diagnosis. medRxiv [Preprint]. 10.1101/2020.10.27.20220541 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

