Microbiome of Field Grown Hemp Reveals Potential Microbial Interactions With Root and Rhizosphere Soil
- PMID: 34867858
- PMCID: PMC8634612
- DOI: 10.3389/fmicb.2021.741597
Microbiome of Field Grown Hemp Reveals Potential Microbial Interactions With Root and Rhizosphere Soil
Abstract
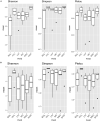
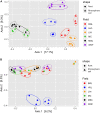
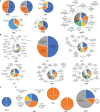
Hemp (Cannabis sativa L.) is a crop bred and grown for the production of fiber, grain, and floral extracts that contribute to health and wellness. Hemp plants interact with a myriad of microbiota inhabiting the phyllosphere, endosphere, rhizoplane, and rhizosphere. These microbes offer many ecological services, particularly those of below ground biotopes which are involved in nutrient cycling, uptake, and alleviating biotic and abiotic stress. The microbiota communities of the hemp rhizosphere in the field are not well documented. To discover core microbiota associated with field grown hemp, we cultivated single C. sativa cultivar, "TJ's CBD," in six different fields in New York and sampled hemp roots and their rhizospheric soil. We used Illumina MiSeq amplicon sequencing targeting 16S ribosomal DNA of bacteria and ITS of fungi to study microbial community structure of hemp roots and rhizospheres. We found that Planctobacteria and Ascomycota dominated the taxonomic composition of hemp associated microbial community. We identified potential core microbiota in each community (bacteria: eight bacterial amplicon sequence variant - ASV, identified as Gimesia maris, Pirellula sp. Lacipirellula limnantheis, Gemmata sp. and unclassified Planctobacteria; fungi: three ASVs identified as Fusarium oxysporum, Gibellulopsis piscis, and Mortierella minutissima). We found 14 ASVs as hub taxa [eight bacterial ASVs (BASV) in the root, and four bacterial and two fungal ASVs in the rhizosphere soil], and 10 BASV connected the root and rhizosphere soil microbiota to form an extended microbial communication in hemp. The only hub taxa detected in both the root and rhizosphere soil microbiota was ASV37 (Caulifigura coniformis), a bacterial taxon. The core microbiota and Network hub taxa can be studied further for biocontrol activities and functional investigations in the formulation of hemp bioinoculants. This study documented the microbial diversity and community structure of hemp grown in six fields, which could contribute toward the development of bioinoculants for hemp that could be used in organic farming.
Keywords: Cannabis sativa; bacterial communities; fungal communities; microbial ecology; microbiome; network analysis; rhizosphere.
Copyright © 2021 Ahmed, Smart and Hijri.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Alabouvette C., Schippers B., Lemanceau P., Bakker P. (1998). “Biological control of fusarium wilts: toward development of commercial products,” in Plant-Microbe Interactions and Biological Control, eds Boland G. J., Kuykendall L. D. (New York: Marcel Deckker; ), 15–38.
LinkOut - more resources
Full Text Sources

