Transcriptome Analysis Reveals Catabolite Control Protein A Regulatory Mechanisms Underlying Glucose-Excess or -Limited Conditions in a Ruminal Bacterium, Streptococcus bovis
- PMID: 34867900
- PMCID: PMC8637274
- DOI: 10.3389/fmicb.2021.767769
Transcriptome Analysis Reveals Catabolite Control Protein A Regulatory Mechanisms Underlying Glucose-Excess or -Limited Conditions in a Ruminal Bacterium, Streptococcus bovis
Abstract
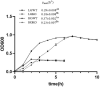
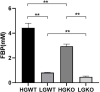
Ruminants may suffer from rumen acidosis when fed with high-concentrate diets due to the higher proliferation and overproduction of lactate by Streptococcus bovis. The catabolite control protein A (CcpA) regulates the transcription of lactate dehydrogenase (ldh) and pyruvate formate-lyase (pfl) in S. bovis, but its role in response to different carbon concentrations remains unclear. To characterize the regulatory mechanisms of CcpA in S. bovis S1 at different levels of carbon, herein, we analyzed the transcriptomic and physiological characteristics of S. bovis S1 and its ccpA mutant strain grown in glucose-excess and glucose-limited conditions. A reduced growth rate and a shift in fermentation pattern from homofermentation to heterofermentation were observed under glucose-limited condition as compared to glucose-excess condition, in S. bovis S1. Additionally, the inactivation of ccpA significantly affected the growth and end metabolites in both conditions. For the glycolytic intermediate, fructose 1,6-bisphosphate (FBP), the concentration significantly reduced at lower glucose conditions; its concentration decreased significantly in the ccpA mutant strain. Transcriptomic results showed that about 46% of the total genes were differentially transcribed between the wild-type strain and ccpA mutant strain grown in glucose-excess conditions; while only 12% genes were differentially transcribed in glucose-limited conditions. Different glucose concentrations led to the differential expression of 38% genes in the wild-type strain, while only half of these were differentially expressed in the ccpA-knockout strain. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses showed that the substrate glucose concentration significantly affected the gene expression in histidine metabolism, nitrogen metabolism, and some carbohydrate metabolism pathways. The deletion of ccpA affected several genes involved in carbohydrate metabolism, such as glycolysis, pyruvate metabolism, fructose and mannose metabolism, as well as in fatty acid biosynthesis pathways in bacteria grown in glucose-excess conditions; this effect was attenuated under glucose-limited conditions. Overall, these findings provide new information on gene transcription and metabolic mechanisms associated with substrate glucose concentration and validate the important role of CcpA in the regulation of carbon metabolism in S. bovis S1 at differential glucose availability.
Keywords: Streptococcus bovis S1; catabolite control protein A; glucose concentration; metabolism regulation; transcriptome.
Copyright © 2021 Jin, Fan, Sun, Zhang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
CcpA-mediated regulation of cellular energy metabolism in the ruminal bacterium Streptococcus bovis.Microbiol Spectr. 2025 Aug 5;13(8):e0215024. doi: 10.1128/spectrum.02150-24. Epub 2025 Jun 25. Microbiol Spectr. 2025. PMID: 40558059 Free PMC article.
-
Regulation of CcpA on the growth and organic acid production characteristics of ruminal Streptococcus bovis at different pH.BMC Microbiol. 2021 Dec 15;21(1):344. doi: 10.1186/s12866-021-02404-x. BMC Microbiol. 2021. PMID: 34911440 Free PMC article.
-
Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis.Appl Environ Microbiol. 2004 Sep;70(9):5244-51. doi: 10.1128/AEM.70.9.5244-5251.2004. Appl Environ Microbiol. 2004. PMID: 15345406 Free PMC article.
-
Carbon catabolite control of the metabolic network in Bacillus subtilis.Biosci Biotechnol Biochem. 2009 Feb;73(2):245-59. doi: 10.1271/bbb.80479. Epub 2009 Feb 7. Biosci Biotechnol Biochem. 2009. PMID: 19202299 Review.
-
Carbon catabolite repression by the catabolite control protein CcpA in Staphylococcus xylosus.J Mol Microbiol Biotechnol. 2002 May;4(3):309-14. J Mol Microbiol Biotechnol. 2002. PMID: 11931563 Review.
Cited by
-
Transcriptional control of carbohydrate catabolism by the CcpA protein in the ruminal bacterium Streptococcus bovis.Appl Environ Microbiol. 2023 Oct 31;89(10):e0047423. doi: 10.1128/aem.00474-23. Epub 2023 Oct 12. Appl Environ Microbiol. 2023. PMID: 37823652 Free PMC article.
-
Transcriptome and Metabolome Analyses Reveal High-Altitude Adaptation Mechanism of Epididymis Sperm Maturation in Tibetan Sheep.Animals (Basel). 2024 Oct 29;14(21):3117. doi: 10.3390/ani14213117. Animals (Basel). 2024. PMID: 39518841 Free PMC article.
-
CcpA-mediated regulation of cellular energy metabolism in the ruminal bacterium Streptococcus bovis.Microbiol Spectr. 2025 Aug 5;13(8):e0215024. doi: 10.1128/spectrum.02150-24. Epub 2025 Jun 25. Microbiol Spectr. 2025. PMID: 40558059 Free PMC article.
-
In Vitro Gene Expression Responses of Bovine Rumen Epithelial Cells to Different pH Stresses.Animals (Basel). 2022 Sep 29;12(19):2621. doi: 10.3390/ani12192621. Animals (Basel). 2022. PMID: 36230362 Free PMC article.
-
Transcriptomic analysis of Lacticaseibacillus paracasei Zhang in transition to the viable but non-culturable state by RNA sequencing.Front Microbiol. 2023 Dec 21;14:1280350. doi: 10.3389/fmicb.2023.1280350. eCollection 2023. Front Microbiol. 2023. PMID: 38188563 Free PMC article.
References
-
- Asanuma N., Hino T. (1997). Tolerance to low pH and lactate production in rumen bacteria. Animal Sci. Technol. 68 367–376.
LinkOut - more resources
Full Text Sources
Miscellaneous

