Raman Imaging of Pathogenic Candida auris: Visualization of Structural Characteristics and Machine-Learning Identification
- PMID: 34867902
- PMCID: PMC8633489
- DOI: 10.3389/fmicb.2021.769597
Raman Imaging of Pathogenic Candida auris: Visualization of Structural Characteristics and Machine-Learning Identification
Abstract
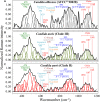
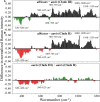
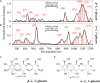
Invasive fungal infections caused by yeasts of the genus Candida carry high morbidity and cause systemic infections with high mortality rate in both immunocompetent and immunosuppressed patients. Resistance rates against antifungal drugs vary among Candida species, the most concerning specie being Candida auris, which exhibits resistance to all major classes of available antifungal drugs. The presently available identification methods for Candida species face a severe trade-off between testing speed and accuracy. Here, we propose and validate a machine-learning approach adapted to Raman spectroscopy as a rapid, precise, and labor-efficient method of clinical microbiology for C. auris identification and drug efficacy assessments. This paper demonstrates that the combination of Raman spectroscopy and machine learning analyses can provide an insightful and flexible mycology diagnostic tool, easily applicable on-site in the clinical environment.
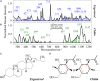
Keywords: Candida auris; Raman imaging; Raman spectroscopy; ergosterol; glucans; machine-learning.
Copyright © 2021 Pezzotti, Kobara, Asai, Nakaya, Miyamoto, Adachi, Yamamoto, Kanamura, Ohgitani, Marin, Zhu, Nishimura, Mazda, Nakata and Makimura.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures