Evolutionary Divergence of the Novel Staphylococcal Species Staphylococcus argenteus
- PMID: 34867903
- PMCID: PMC8640356
- DOI: 10.3389/fmicb.2021.769642
Evolutionary Divergence of the Novel Staphylococcal Species Staphylococcus argenteus
Abstract
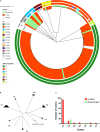
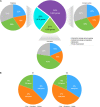
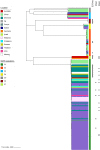
Currently, invasive infections caused by Staphylococcus argenteus, which is a recently named staphylococcal species, are increasingly reported worldwide. However, only a few genomic studies of S. argenteus have offered comprehensive information regarding its genetic diversity, epidemiological characteristics, antimicrobial resistance genes (ARGs), virulence genes and other profiles. Here, we describe a comparative genomic analysis by population structure, pangenome, panmobilome, region-specific accessory genes confer an adaptive advantage in 153 S. argenteus strains which comprised 24 strains sequenced in this study and 129 strains whose genome sequences were available from GenBank. As a result, the population of S. argenteus comprised seven genetically distinct clades, including two major clades (C1 and C2), with distinct isolation source patterns. Pangenome analysis revealed that S. argenteus has an open pangenome composed of 7,319 genes and a core genome composed of 1,508 genes. We further determined the distributions of 75 virulence factors (VFs) and 30 known ARGs and identified at least four types of plasmids and 93 complete or partial putative prophages. It indicate that S. argenteus may show a similar level of pathogenicity to that of S. aureus. This study also provides insights into the evolutionary divergence of this pathogen, indicating that the geographical distribution was a potential driving force behind the evolutionary divergence of S. argenteus. The preferential horizontal acquisition of particular elements, such as staphylococcal cassette chromosome mec elements and plasmids, was observed in specific regions, revealing potential gene exchange between S. argenteus strains and local S. aureus strains. Moreover, multiple specific genes related to environmental adaptation were identified in strains isolated from East Asia. However, these findings may help promote our understanding of the evolutionary divergence of this bacterium at a high genetic resolution by providing insights into the epidemiology of S. argenteus and may help combat its spread.
Keywords: Staphylococcus argenteus; genetic divergence; pangenome; population structure; regional specialization.
Copyright © 2021 Wu, Pang, Huang, Zhang, Cai, Zhang, Chen, Xue, Gu, Wang, Ding, Wan and Wu.
Conflict of interest statement
ZC and QWa are employed by Guangdong Huankai Microbial Science and Technology Co. Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Preliminary comparative genomics revealed pathogenic potential and international spread of Staphylococcus argenteus.BMC Genomics. 2017 Oct 23;18(1):808. doi: 10.1186/s12864-017-4149-9. BMC Genomics. 2017. PMID: 29058585 Free PMC article.
-
Molecular characterization of Staphylococcus argenteus in Myanmar: identification of novel genotypes/clusters in staphylocoagulase, protein A, alpha-haemolysin and other virulence factors.J Med Microbiol. 2019 Jan;68(1):95-104. doi: 10.1099/jmm.0.000869. Epub 2018 Nov 12. J Med Microbiol. 2019. PMID: 30418108
-
Prevalence and characterization of Staphylococcus aureus and Staphylococcus argenteus isolated from rice and flour products in Guangdong, China.Int J Food Microbiol. 2023 Dec 2;406:110348. doi: 10.1016/j.ijfoodmicro.2023.110348. Epub 2023 Aug 6. Int J Food Microbiol. 2023. PMID: 37573713
-
Whole Genome Sequencing of Danish Staphylococcus argenteus Reveals a Genetically Diverse Collection with Clear Separation from Staphylococcus aureus.Front Microbiol. 2017 Aug 9;8:1512. doi: 10.3389/fmicb.2017.01512. eCollection 2017. Front Microbiol. 2017. PMID: 28848522 Free PMC article.
-
Description of Staphylococcal Strains from Straw-Coloured Fruit Bat (Eidolon helvum) and Diamond Firetail (Stagonopleura guttata) and a Review of their Phylogenetic Relationships to Other Staphylococci.Front Cell Infect Microbiol. 2022 May 11;12:878137. doi: 10.3389/fcimb.2022.878137. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35646742 Free PMC article. Review.
Cited by
-
The Staphylococcus aureus complex: implications for the clinical microbiology laboratory.J Clin Microbiol. 2025 Jul 9;63(7):e0127624. doi: 10.1128/jcm.01276-24. Epub 2025 Jun 4. J Clin Microbiol. 2025. PMID: 40464548 Free PMC article. Review.
-
Isolation and Genomic Analysis of a Case of Staphylococcus argenteus ST2250 Related to Sepsis in Italy.Microorganisms. 2024 Jul 20;12(7):1485. doi: 10.3390/microorganisms12071485. Microorganisms. 2024. PMID: 39065252 Free PMC article.
-
Prevalence and Characteristics of Invasive Staphylococcus argenteus among Patients with Bacteremia in Hong Kong.Microorganisms. 2023 Sep 28;11(10):2435. doi: 10.3390/microorganisms11102435. Microorganisms. 2023. PMID: 37894094 Free PMC article.
-
Investigation of methicillin-resistant Staphylococcus aureus, methicillin-susceptible Staphylococcus aureus, and Staphylococcus argenteus from wild long-tailed macaques (Macaca fascicularis) at Kosumpee Forest Park, Maha Sarakham, Thailand.Vet World. 2022 Nov;15(11):2693-2698. doi: 10.14202/vetworld.2022.2693-2698. Epub 2022 Nov 26. Vet World. 2022. PMID: 36590126 Free PMC article.
-
Genomic analysis and identification of a novel superantigen, SargEY, in Staphylococcus argenteus isolated from atopic dermatitis lesions.mSphere. 2024 Jul 30;9(7):e0050524. doi: 10.1128/msphere.00505-24. Epub 2024 Jul 11. mSphere. 2024. PMID: 38990001 Free PMC article.
References
-
- Argudín M. A., Dodémont M., Vandendriessche S., Rottiers S., Tribes C., Roisin S., et al. (2016). Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. Eur. J. Clin. Microbiol. Infect. Dis. 35 1017–1022. 10.1007/s10096-016-2632-x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

