Comparative Genomic and Pan-Genomic Characterization of Staphylococcus epidermidis From Different Sources Unveils the Molecular Basis and Potential Biomarkers of Pathogenic Strains
- PMID: 34867904
- PMCID: PMC8634615
- DOI: 10.3389/fmicb.2021.770191
Comparative Genomic and Pan-Genomic Characterization of Staphylococcus epidermidis From Different Sources Unveils the Molecular Basis and Potential Biomarkers of Pathogenic Strains
Abstract
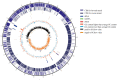
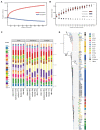
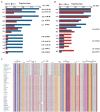
Coagulase-negative Staphylococcus (CoNS) is the most common pathogen causing traumatic endophthalmitis. Among which, Staphylococcus epidermidis is the most common species that colonizes human skin, eye surfaces, and nasal cavity. It is also the main cause of nosocomial infection, specially foreign body-related bloodstream infections (FBR-BSIs). Although some studies have reported the genome characteristics of S. epidermidis, the genome of ocular trauma-sourced S. epidermidis strain and a comprehensive understanding of its pathogenicity are still lacking. Our study sequenced, analyzed, and reported the whole genomes of 11 ocular trauma-sourced samples of S. epidermidis that caused traumatic endophthalmitis. By integrating publicly available genomes, we obtained a total of 187 S. epidermidis samples from healthy and diseased eyes, skin, respiratory tract, and blood. Combined with pan-genome, phylogenetic, and comparative genomic analyses, our study showed that S. epidermidis, regardless of niche source, exhibits two founder lineages with different pathogenicity. Moreover, we identified several potential biomarkers associated with the virulence of S. epidermidis, including essD, uhpt, sdrF, sdrG, fbe, and icaABCDR. EssD and uhpt have high homology with esaD and hpt in Staphylococcus aureus, showing that the genomes of S. epidermidis and S. aureus may have communicated during evolution. SdrF, sdrG, fbe, and icaABCDR are related to biofilm formation. Compared to S. epidermidis from blood sources, ocular-sourced strains causing intraocular infection had no direct relationship with biofilm formation. In conclusion, this study provided additional data resources for studies on S. epidermidis and improved our understanding of the evolution and pathogenicity among strains of different sources.
Keywords: Staphylococcus epidermidis; antimicrobial resistance; comparative genomic; evolution; ocular; pan-genomic; virulence gene; whole genome.
Copyright © 2021 Lin, Sun, Shi, Xu, Gu, Gu, Ma, Wan, Xu, Su, Lou and Zheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Comparative virulome analysis of four Staphylococcus epidermidis strains from human skin and platelet concentrates using whole genome sequencing.Access Microbiol. 2024 Apr 3;6(4):000780.v3. doi: 10.1099/acmi.0.000780.v3. eCollection 2024. Access Microbiol. 2024. PMID: 38737800 Free PMC article.
-
Comparative Genome Analysis Reveals the Molecular Basis of Niche Adaptation of Staphylococcus epidermidis Strains.Front Genet. 2020 Nov 9;11:566080. doi: 10.3389/fgene.2020.566080. eCollection 2020. Front Genet. 2020. PMID: 33240320 Free PMC article.
-
Staphylococcus epidermidis Phages Transduce Antimicrobial Resistance Plasmids and Mobilize Chromosomal Islands.mSphere. 2021 May 12;6(3):e00223-21. doi: 10.1128/mSphere.00223-21. mSphere. 2021. PMID: 33980677 Free PMC article.
-
Host Response to Staphylococcus epidermidis Colonization and Infections.Front Cell Infect Microbiol. 2017 Mar 21;7:90. doi: 10.3389/fcimb.2017.00090. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28377905 Free PMC article. Review.
-
Phenotypic characteristics of coagulase-negative staphylococci: typing and antibiotic susceptibility.APMIS Suppl. 1999;91:1-42. APMIS Suppl. 1999. PMID: 10230367 Review.
Cited by
-
Beyond the Wild MRSA: Genetic Features and Phylogenomic Review of mecC-Mediated Methicillin Resistance in Non-aureus Staphylococci and Mammaliicocci.Microorganisms. 2023 Dec 29;12(1):66. doi: 10.3390/microorganisms12010066. Microorganisms. 2023. PMID: 38257893 Free PMC article. Review.
-
[Distribution and Antibiotic Resistance Analysis of Ocular Bacterial Pathogens at a Tertiary Hospital From 2012 to 2021].Sichuan Da Xue Xue Bao Yi Xue Ban. 2024 Jan 20;55(1):204-209. doi: 10.12182/20240160103. Sichuan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38322538 Free PMC article. Chinese.
-
Difficult-to-Treat Pathogens: A Review on the Management of Multidrug-Resistant Staphylococcus epidermidis.Life (Basel). 2023 May 4;13(5):1126. doi: 10.3390/life13051126. Life (Basel). 2023. PMID: 37240771 Free PMC article. Review.
-
The impact of oxygen content on Staphylococcus epidermidis pathogenesis in ocular infection based on clinical characteristics, transcriptome and metabolome analysis.Front Microbiol. 2024 Jul 10;15:1409597. doi: 10.3389/fmicb.2024.1409597. eCollection 2024. Front Microbiol. 2024. PMID: 39050640 Free PMC article.
-
Genomic analysis of multi-drug resistant coagulase-negative staphylococci from healthy humans and animals revealed unusual mechanisms of resistance and CRISPR-Cas system.Int Microbiol. 2025 Jun;28(5):941-963. doi: 10.1007/s10123-024-00577-9. Epub 2024 Sep 17. Int Microbiol. 2025. PMID: 39287832 Free PMC article.
References
-
- Al-Omran A. M., Abboud E. B., Abu El-Asrar A. M. (2007). Microbiologic spectrum and visual outcome of posttraumatic endophthalmitis. Retina 27, 236–242. doi: 10.1097/01.iae.0000225072.68265.ee, PMID: - DOI - PubMed
-
- Asbell P. A., Sahm D. F., Shaw M., Draghi D. C., Brown N. P. (2008). Increasing prevalence of methicillin resistance in serious ocular infections caused by Staphylococcus aureus in the United States: 2000 to 2005. J. Cataract Refract. Surg. 34, 814–818. doi: 10.1016/j.jcrs.2008.01.016, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

