Comparative Proteomics Demonstrates Altered Metabolism Pathways in Cotrimoxazole- Resistant and Amikacin-Resistant Klebsiella pneumoniae Isolates
- PMID: 34867912
- PMCID: PMC8637018
- DOI: 10.3389/fmicb.2021.773829
Comparative Proteomics Demonstrates Altered Metabolism Pathways in Cotrimoxazole- Resistant and Amikacin-Resistant Klebsiella pneumoniae Isolates
Abstract
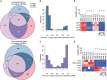
Antibiotic resistance (AMR) has always been a hot topic all over the world and its mechanisms are varied and complicated. Previous evidence revealed the metabolic slowdown in resistant bacteria, suggesting the important role of metabolism in antibiotic resistance. However, the molecular mechanism of reduced metabolism remains poorly understood, which inspires us to explore the global proteome change during antibiotic resistance. Here, the sensitive, cotrimoxazole-resistant, amikacin-resistant, and amikacin/cotrimoxazole -both-resistant KPN clinical isolates were collected and subjected to proteome analysis through liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). A deep coverage of 2,266 proteins were successfully identified and quantified in total, representing the most comprehensive protein quantification data by now. Further bioinformatic analysis showed down-regulation of tricarboxylic acid cycle (TCA) pathway and up-regulation of alcohol metabolic or glutathione metabolism processes, which may contribute to ROS clearance and cell survival, in drug-resistant isolates. These results indicated that metabolic pathway alteration was directly correlated with antibiotic resistance, which could promote the development of antibacterial drugs from "target" to "network." Moreover, combined with minimum inhibitory concentration (MIC) of cotrimoxazole and amikacin on different KPN isolates, we identified nine proteins, including garK, uxaC, exuT, hpaB, fhuA, KPN_01492, fumA, hisC, and aroE, which might contribute mostly to the survival of KPN under drug pressure. In sum, our findings provided novel, non-antibiotic-based therapeutics against resistant KPN.
Keywords: Klebsiella pneumoniae (K. pneumoniae); antibiotic resistance; bioinformatics; comparative proteomics; metabolism.
Copyright © 2021 Shen, Shen, Zhang, Xu, He and Qie.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

