Data-Driven Analysis of COVID-19 Reveals Persistent Immune Abnormalities in Convalescent Severe Individuals
- PMID: 34867943
- PMCID: PMC8640498
- DOI: 10.3389/fimmu.2021.710217
Data-Driven Analysis of COVID-19 Reveals Persistent Immune Abnormalities in Convalescent Severe Individuals
Abstract
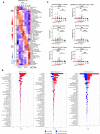
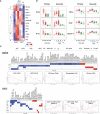
Severe SARS-CoV-2 infection can trigger uncontrolled innate and adaptive immune responses, which are commonly associated with lymphopenia and increased neutrophil counts. However, whether the immune abnormalities observed in mild to severely infected patients persist into convalescence remains unclear. Herein, comparisons were drawn between the immune responses of COVID-19 infected and convalescent adults. Strikingly, survivors of severe COVID-19 had decreased proportions of NKT and Vδ2 T cells, and increased proportions of low-density neutrophils, IgA+/CD86+/CD123+ non-classical monocytes and hyperactivated HLADR+CD38+ CD8+ T cells, and elevated levels of pro-inflammatory cytokines such as hepatocyte growth factor and vascular endothelial growth factor A, long after virus clearance. Our study suggests potential immune correlates of "long COVID-19", and defines key cells and cytokines that delineate true and quasi-convalescent states.
Keywords: COVID-19; SARS – CoV – 2; active infection; cytokine profile; immune recovery; immunophenotyping; inflammation; severity.
Copyright © 2021 Lim, Puan, Wang, Teng, Loh, Tan, Carissimo, Chan, Poh, Lee, Fong, Yeo, Chee, Amrun, Chang, Tay, Torres-Ruesta, Leo Fernandez, How, Andiappan, Lee, Duan, Tan, Yan, Kalimuddin, Lye, Leo, Ong, Young, Renia, Ng, Lee and Rötzschke.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Jouan Y, Guillon A, Gonzalez L, Perez Y, Ehrmann S, Ferreira M, et al. Functional Alteration of Innate T Cells in Critically Ill Covid-19 Patients. medRxiv (2020) 2020. doi: 10.1101/2020.05.03.20089300. 05.03.20089300. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

