SSTP1, a Host Defense Peptide, Exploits the Immunomodulatory IL6 Pathway to Induce Apoptosis in Cancer Cells
- PMID: 34867962
- PMCID: PMC8639500
- DOI: 10.3389/fimmu.2021.740620
SSTP1, a Host Defense Peptide, Exploits the Immunomodulatory IL6 Pathway to Induce Apoptosis in Cancer Cells
Abstract
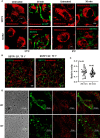
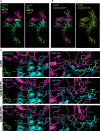
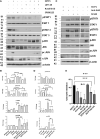
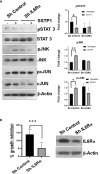
While the immunomodulatory pathways initiated in immune cells contribute to therapeutic response, their activation in cancer cells play a role in cancer progression. Also, many of the aberrantly expressed immunomodulators on cancer cells are considered as therapeutic targets. Here, we introduce host defense peptide (HDP), a known immuomodulator, as a therapeutic agent to target them. The cationic host defense peptides (HDPs), an integral part of the innate immune system, possess membranolytic activity, which imparts antimicrobial and antitumor efficacy to it. They act as immunomodulators by activating the immune cells. Though their antimicrobial function has been recently reassigned to immunoregulation, their antitumor activity is still attributed to its membranolytic activity. This membrane pore formation ability, which is proportional to the concentration of the peptide, also leads to side effects like hemolysis, limiting their therapeutic application. So, despite the identification of a variety of anticancer HDPs, their clinical utility is limited. Though HDPs are shown to exert the immunomodulatory activity through specific membrane targets on immune cells, their targets on cancer cells are unknown. We show that SSTP1, a novel HDP identified by shotgun cloning, binds to the active IL6/IL6Rα/gp130 complex on cancer cells, rearranging the active site residues. In contrast to the IL6 blockers inhibiting JAK/STAT activity, SSTP1 shifts the proliferative IL6/JAK/STAT signaling to the apoptotic IL6/JNK/AP1 pathway. In IL6Rα-overexpressing cancer cells, SSTP1 induces apoptosis at low concentration through JNK pathway, without causing significant membrane disruption. We highlight the importance of immunomodulatory pathways in cancer apoptosis, apart from its established role in immune cell regulation and cancer cell proliferation. Our study suggests that identification of the membrane targets for the promising anticancer HDPs might lead to the identification of new drugs for targeted therapy.
Keywords: IL6 (IL-6) pathway; JNK/AP1 pathway; apoptosis; cancer; host defense peptides; immunoregulation; temporins.
Copyright © 2021 Gopalakrishnan, Uma, Mohan, Mohan, Shanmugam, Kumar, J, Chandrika, Vasudevan, Nori, Sathi, George and Maliekal.
Conflict of interest statement
A patent application has been filed for the peptide, SSTP1 in USA. “Apoptosis Inducing Peptide (SSTP1)”, PCT/IN2021/050371, in the name of Rajiv Gandhi Centre for Biotechnology, with TM & SG as Investigators. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

