Acute and Chronic Changes in Gene Expression After CMV DNAemia in Kidney Transplant Recipients
- PMID: 34867983
- PMCID: PMC8634678
- DOI: 10.3389/fimmu.2021.750659
Acute and Chronic Changes in Gene Expression After CMV DNAemia in Kidney Transplant Recipients
Abstract
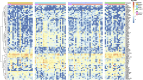
Cytomegalovirus (CMV) viremia continues to cause significant morbidity and mortality in kidney transplant patients with clinical complications including organ rejection and death. Whole blood gene expression dynamics in CMV viremic patients from onset of DNAemia through convalescence has not been well studied to date in humans. To evaluate how CMV infection impacts whole blood leukocyte gene expression over time, we evaluated a matched cohort of 62 kidney transplant recipients with and without CMV DNAemia using blood samples collected at multiple time points during the 12-month period after transplant. While transcriptomic differences were minimal at baseline between DNAemic and non-DNAemic patients, hundreds of genes were differentially expressed at the long-term timepoint, including genes enriching for pathways important for macrophages, interferon, and IL-8 signaling. Amongst patients with CMV DNAemia, the greatest amount of transcriptomic change occurred between baseline and 1-week post-DNAemia, with increase in pathways for interferon signaling and cytotoxic T cell function. Time-course gene set analysis of these differentially expressed genes revealed that most of the enriched pathways had a significant time-trend. While many pathways that were significantly down- or upregulated at 1 week returned to baseline-like levels, we noted that several pathways important in adaptive and innate cell function remained upregulated at the long-term timepoint after resolution of CMV DNAemia. Differential expression analysis and time-course gene set analysis revealed the dynamics of genes and pathways involved in the immune response to CMV DNAemia in kidney transplant patients. Understanding transcriptional changes caused by CMV DNAemia may identify the mechanism behind patient vulnerability to CMV reactivation and increased risk of rejection in transplant recipients and suggest protective strategies to counter the negative immunologic impact of CMV. These findings provide a framework to identify immune correlates for risk assessment and guiding need for extending antiviral prophylaxis.
Keywords: CMV DNAemia; RNA-seq; kidney transplant; transcriptomics; transplant immunology.
Copyright © 2021 Ahn, Schaenman, Qian, Pickering, Groysberg, Rossetti, Llamas, Hoffmann, Gjertson, Deng, Bunnapradist, Reed and CMV Systems Immunobiology Group.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

