The Transcriptome and Metabolome Reveal Stress Responses in Sulfur-Fumigated Cucumber (Cucumis sativus L.)
- PMID: 34868181
- PMCID: PMC8636124
- DOI: 10.3389/fpls.2021.778956
The Transcriptome and Metabolome Reveal Stress Responses in Sulfur-Fumigated Cucumber (Cucumis sativus L.)
Abstract
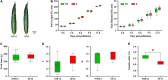
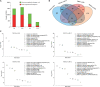
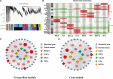
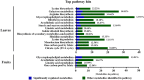
Sulfur (S) fumigation is a commonly used sterilization method in horticultural facilities against fungal diseases. S fumigation damaged cucumber leaves, although the response mechanism is unclear. This study analyzes the growth, transcriptome, and metabolomic profiles of young and mature leaves, ovaries, and commercial cucumber fruits to decipher the mechanism of cucumber stress response under S fumigation. S fumigation significantly changed the photosynthetic efficiency and reactive oxygen species (ROS) in leaves, but not fruit development, fruit mass, and peel color. Transcriptome analysis indicated that S fumigation strongly regulated stress defense genes. The weighted gene co-expression network analysis revealed that S fumigation regulated ASPG1, AMC1 defense genes, LECRK3, and PERK1 protein kinase. The abscisic acid (ABA)-mediated model of regulation under S fumigation was constructed. Metabolome analysis showed that S fumigation significantly upregulated or downregulated the contents of amino acids, organic acids, sugars, glycosides, and lipids (VIP > 1 and P-value < 0.05). The opposite Pearson's correlations of these differential metabolites implied that cucumber had different metabolic patterns in short-term and long-term S fumigation. Besides, the elevated levels of proline and triglyceride indicated that stress-responsive mechanisms existed in S-fumigated cucumber. Moreover, the comprehensive analysis indicated that S fumigation elevated secondary S-containing metabolites but decreased sulfate absorption and transportation in cucumber. Overall, our results provided a comprehensive assessment of S fumigation on cucumber, which laid the theoretical foundation for S fumigation in protected cultivation.
Keywords: cucumber; metabolome; stress response; sulfur fumigation; transcriptome.
Copyright © 2021 Liu, Gao, Gong, Hou, Zhang, Cheng, Du, Zhang, Wang, Xu, Xing, Kang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Integrated Metabolome and Transcriptome Analysis Provide Insights into the Effects of Grafting on Fruit Flavor of Cucumber with Different Rootstocks.Int J Mol Sci. 2019 Jul 23;20(14):3592. doi: 10.3390/ijms20143592. Int J Mol Sci. 2019. PMID: 31340498 Free PMC article.
-
Integration of transcriptome and metabolome reveals key regulatory defense pathways associated with high temperature stress in cucumber (Cucumis sativus L.).BMC Plant Biol. 2025 Jan 2;25(1):6. doi: 10.1186/s12870-024-05876-x. BMC Plant Biol. 2025. PMID: 39748295 Free PMC article.
-
Integrated Metabolome and Transcriptome Analysis Unveils Novel Pathway Involved in the Formation of Yellow Peel in Cucumber.Int J Mol Sci. 2021 Feb 2;22(3):1494. doi: 10.3390/ijms22031494. Int J Mol Sci. 2021. PMID: 33540857 Free PMC article.
-
Metabolome and transcriptome analyses reveal chlorophyll and anthocyanin metabolism pathway associated with cucumber fruit skin color.BMC Plant Biol. 2020 Aug 24;20(1):386. doi: 10.1186/s12870-020-02597-9. BMC Plant Biol. 2020. PMID: 32831013 Free PMC article.
-
[Effects of lime-ammonium bicarbonate fumigation and biofertilizer application on Fusarium wilt and biomass of continuous cropping cucumber and watermelon.].Ying Yong Sheng Tai Xue Bao. 2017 Oct;28(10):3351-3359. doi: 10.13287/j.1001-9332.201710.036. Ying Yong Sheng Tai Xue Bao. 2017. PMID: 29692155 Chinese.
Cited by
-
Multiple Stressors in Vegetable Production: Insights for Trait-Based Crop Improvement in Cucurbits.Front Plant Sci. 2022 May 3;13:861637. doi: 10.3389/fpls.2022.861637. eCollection 2022. Front Plant Sci. 2022. PMID: 35592574 Free PMC article. Review.
-
Organic acid metabolism in Chinese dwarf cherry [Cerasus humilis (Bge.) Sok.] is controlled by a complex gene regulatory network.Front Plant Sci. 2022 Sep 2;13:982112. doi: 10.3389/fpls.2022.982112. eCollection 2022. Front Plant Sci. 2022. PMID: 36160985 Free PMC article.
-
Responses of differential metabolites and pathways to high temperature in cucumber anther.Front Plant Sci. 2023 Apr 14;14:1131735. doi: 10.3389/fpls.2023.1131735. eCollection 2023. Front Plant Sci. 2023. PMID: 37123826 Free PMC article.
-
Metabolomics-Driven Mining of Metabolite Resources: Applications and Prospects for Improving Vegetable Crops.Int J Mol Sci. 2022 Oct 11;23(20):12062. doi: 10.3390/ijms232012062. Int J Mol Sci. 2022. PMID: 36292920 Free PMC article. Review.
References
-
- Anders S., Huber W. (2013). Differential expression of RNA-Seq data at the gene level - the DESeq package. EMBL.
-
- Auler P. A., Souza G. M., da Silva, Engela M. R. G., do Amaral M. N., Rossatto T., et al. (2021). Stress memory of physiological, biochemical and metabolomic responses in two different rice genotypes under drought stress: The scale matters. Plant Sci. 311:110994. 10.1016/J.PLANTSCI.2021.110994 - DOI - PubMed
LinkOut - more resources
Full Text Sources

