Transcriptome Profiling of the Salt Stress Response in the Leaves and Roots of Halophytic Eutrema salsugineum
- PMID: 34868259
- PMCID: PMC8637539
- DOI: 10.3389/fgene.2021.770742
Transcriptome Profiling of the Salt Stress Response in the Leaves and Roots of Halophytic Eutrema salsugineum
Abstract
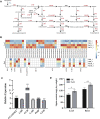
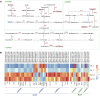
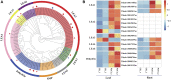
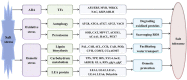
Eutrema salsugineum can grow in natural harsh environments; however, the underlying mechanisms for salt tolerance of Eutrema need to be further understood. Herein, the transcriptome profiling of Eutrema leaves and roots exposed to 300 mM NaCl is investigated, and the result emphasized the role of genes involved in lignin biosynthesis, autophagy, peroxisome, and sugar metabolism upon salt stress. Furthermore, the expression of the lignin biosynthesis and autophagy-related genes, as well as 16 random selected genes, was validated by qRT-PCR. Notably, the transcript abundance of a large number of lignin biosynthesis genes such as CCoAOMT, C4H, CCR, CAD, POD, and C3'H in leaves was markedly elevated by salt shock. And the examined lignin content in leaves and roots demonstrated salt stress led to lignin accumulation, which indicated the enhanced lignin level could be an important mechanism for Eutrema responding to salt stress. Additionally, the differentially expressed genes (DEGs) assigned in the autophagy pathway including Vac8, Atg8, and Atg4, as well as DEGs enriched in the peroxisome pathway such as EsPEX7, EsCAT, and EsSOD2, were markedly induced in leaves and/or roots. In sugar metabolism pathways, the transcript levels of most DEGs associated with the synthesis of sucrose, trehalose, raffinose, and xylose were significantly enhanced. Furthermore, the expression of various stress-related transcription factor genes including WRKY, AP2/ERF-ERF, NAC, bZIP, MYB, C2H2, and HSF was strikingly improved. Collectively, the increased expression of biosynthesis genes of lignin and soluble sugars, as well as the genes in the autophagy and peroxisome pathways, suggested that Eutrema encountering salt shock possibly possess a higher capacity to adjust osmotically and facilitate water transport and scavenge reactive oxidative species and oxidative proteins to cope with the salt environment. Thus, this study provides a new insight for exploring the salt tolerance mechanism of halophytic Eutrema and discovering new gene targets for the genetic improvement of crops.
Keywords: Eutrema salsugineum; RNA-seq; autophagy; lignin biosynthesis; peroxisome; salt shock; sugar metabolism; transcription factor.
Copyright © 2021 Li, Qi, Zhao, Wang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

