Identification of Hub Genes Correlated With Poor Prognosis for Patients With Uterine Corpus Endometrial Carcinoma by Integrated Bioinformatics Analysis and Experimental Validation
- PMID: 34868993
- PMCID: PMC8639584
- DOI: 10.3389/fonc.2021.766947
Identification of Hub Genes Correlated With Poor Prognosis for Patients With Uterine Corpus Endometrial Carcinoma by Integrated Bioinformatics Analysis and Experimental Validation
Abstract
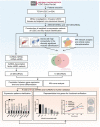
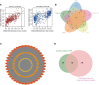
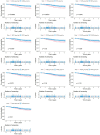
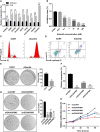
Uterine Corpus Endometrial Carcinoma (UCEC) is one of the most common malignancies of the female genital tract and there remains a major public health problem. Although significant progress has been made in explaining the progression of UCEC, it is still warranted that molecular mechanisms underlying the tumorigenesis of UCEC are to be elucidated. The aim of the current study was to investigate key modules and hub genes related to UCEC pathogenesis, and to explore potential biomarkers and therapeutic targets for UCEC. The RNA-seq dataset and corresponding clinical information for UCEC patients were obtained from the Cancer Genome Atlas (TCGA) database. Differentially expressed genes (DEGs) were screened between 23 paired UCEC tissues and adjacent non-cancerous tissues. Subsequently, the co-expression network of DEGs was determined via weighted gene co-expression network analysis (WGCNA). The Blue and Brown modules were identified to be significantly positively associated with neoplasm histologic grade. The highly connected genes of the two modules were then investigated as potential key factors related to tumor differentiation. Additionally, a protein-protein interaction (PPI) network for all genes in the two modules was constructed to obtain key modules and nodes. 10 genes were identified by both WGCNA and PPI analyses, and it was shown by Kaplan-Meier curve analysis that 6 out of the 10 genes were significantly negatively related to the 5-year overall survival (OS) in patients (AURKA, BUB1, CDCA8, DLGAP5, KIF2C, TPX2). Besides, according to the DEGs from the two modules, lncRNA-miRNA-mRNA and lncRNA-TF-mRNA networks were constructed to explore the molecular mechanism of UCEC-related lncRNAs. 3 lncRNAs were identified as being significantly negatively related to the 5-year OS (AC015849.16, DUXAP8 and DGCR5), with higher expression in UCEC tissues compared to non-tumor tissues. Finally, quantitative Real-time PCR was applied to validate the expression patterns of hub genes. Cell proliferation and colony formation assays, as well as cell cycle distribution and apoptosis analysis, were performed to test the effects of representative hub genes. Altogether, this study not only promotes our understanding of the molecular mechanisms for the pathogenesis of UCEC but also identifies several promising biomarkers in UCEC development, providing potential therapeutic targets for UCEC.
Keywords: hub gene; protein-protein interaction network; tumor differentiation; uterine corpus endometrial carcinoma; weighted gene co-expression network analysis.
Copyright © 2021 Yuan, Chen, Cai, He, Li and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

