Can the SARS-CoV-2 Spike Protein Bind Integrins Independent of the RGD Sequence?
- PMID: 34869067
- PMCID: PMC8637727
- DOI: 10.3389/fcimb.2021.765300
Can the SARS-CoV-2 Spike Protein Bind Integrins Independent of the RGD Sequence?
Abstract
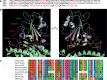
The RGD motif in the Severe Acute Syndrome Coronavirus 2 (SARS-CoV-2) spike protein has been predicted to bind RGD-recognizing integrins. Recent studies have shown that the spike protein does, indeed, interact with αVβ3 and α5β1 integrins, both of which bind to RGD-containing ligands. However, computational studies have suggested that binding between the spike RGD motif and integrins is not favourable, even when unfolding occurs after conformational changes induced by binding to the canonical host entry receptor, angiotensin-converting enzyme 2 (ACE2). Furthermore, non-RGD-binding integrins, such as αx, have been suggested to interact with the SARS-CoV-2 spike protein. Other viral pathogens, such as rotaviruses, have been recorded to bind integrins in an RGD-independent manner to initiate host cell entry. Thus, in order to consider the potential for the SARS-CoV-2 spike protein to bind integrins independent of the RGD sequence, we investigate several factors related to the involvement of integrins in SARS-CoV-2 infection. First, we review changes in integrin expression during SARS-CoV-2 infection to identify which integrins might be of interest. Then, all known non-RGD integrin-binding motifs are collected and mapped to the spike protein receptor-binding domain and analyzed for their 3D availability. Several integrin-binding motifs are shown to exhibit high sequence similarity with solvent accessible regions of the spike receptor-binding domain. Comparisons of these motifs with other betacoronavirus spike proteins, such as SARS-CoV and RaTG13, reveal that some have recently evolved while others are more conserved throughout phylogenetically similar betacoronaviruses. Interestingly, all of the potential integrin-binding motifs, including the RGD sequence, are conserved in one of the known pangolin coronavirus strains. Of note, the most recently recorded mutations in the spike protein receptor-binding domain were found outside of the putative integrin-binding sequences, although several mutations formed inside and close to one motif, in particular, may potentially enhance binding. These data suggest that the SARS-CoV-2 spike protein may interact with integrins independent of the RGD sequence and may help further explain how SARS-CoV-2 and other viruses can evolve to bind to integrins.
Keywords: RGD; SARS-CoV-2; SARS-CoV-2 spike protein; bioinformatics; integrin; integrin-binding motif.
Copyright © 2021 Beaudoin, Hamaia, Huang, Blundell and Jackson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aguirre C., Meca-Lallana V., Barrios-Blandino A., Del Río B., Vivancos J. (2020). Covid-19 in a Patient With Multiple Sclerosis Treated With Natalizumab: May the Blockade of Integrins Have a Protective Role? Mult. Scler. Relat. Disord. 44, 102250. doi: 10.1016/j.msard.2020.102250 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

