Zyxin Is Involved in Fibroblast Rigidity Sensing and Durotaxis
- PMID: 34869319
- PMCID: PMC8637444
- DOI: 10.3389/fcell.2021.735298
Zyxin Is Involved in Fibroblast Rigidity Sensing and Durotaxis
Abstract
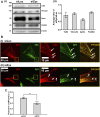
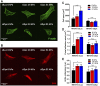
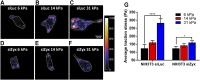
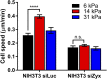
Focal adhesions (FAs) are specialized structures that enable cells to sense their extracellular matrix rigidity and transmit these signals to the interior of the cells, bringing about actin cytoskeleton reorganization, FA maturation, and cell migration. It is known that cells migrate towards regions of higher substrate rigidity, a phenomenon known as durotaxis. However, the underlying molecular mechanism of durotaxis and how different proteins in the FA are involved remain unclear. Zyxin is a component of the FA that has been implicated in connecting the actin cytoskeleton to the FA. We have found that knocking down zyxin impaired NIH3T3 fibroblast's ability to sense and respond to changes in extracellular matrix in terms of their FA sizes, cell traction stress magnitudes and F-actin organization. Cell migration speed of zyxin knockdown fibroblasts was also independent of the underlying substrate rigidity, unlike wild type fibroblasts which migrated fastest at an intermediate substrate rigidity of 14 kPa. Wild type fibroblasts exhibited durotaxis by migrating toward regions of increasing substrate rigidity on polyacrylamide gels with substrate rigidity gradient, while zyxin knockdown fibroblasts did not exhibit durotaxis. Therefore, we propose zyxin as an essential protein that is required for rigidity sensing and durotaxis through modulating FA sizes, cell traction stress and F-actin organization.
Keywords: durotaxis; focal adhesion; mechanotransduction; rigidity sensing; zyxin.
Copyright © 2021 Yip, Zhang, Chong, Cheruba, Woon, Chua, Goh, Yang, Tay, Koh and Chiam.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Substrates with engineered step changes in rigidity induce traction force polarity and durotaxis.Cell Mol Bioeng. 2014 Mar;7(1):26-34. doi: 10.1007/s12195-013-0307-6. Epub 2013 Oct 9. Cell Mol Bioeng. 2014. PMID: 27721906 Free PMC article.
-
The focal adhesion-localized CdGAP regulates matrix rigidity sensing and durotaxis.PLoS One. 2014 Mar 14;9(3):e91815. doi: 10.1371/journal.pone.0091815. eCollection 2014. PLoS One. 2014. PMID: 24632816 Free PMC article.
-
Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration.Cell. 2012 Dec 21;151(7):1513-27. doi: 10.1016/j.cell.2012.11.034. Cell. 2012. PMID: 23260139 Free PMC article.
-
Positive and negative durotaxis - mechanisms and emerging concepts.J Cell Sci. 2024 Apr 15;137(8):jcs261919. doi: 10.1242/jcs.261919. Epub 2024 Apr 22. J Cell Sci. 2024. PMID: 38647525 Review.
-
Guiding cell migration by tugging.Curr Opin Cell Biol. 2013 Oct;25(5):619-26. doi: 10.1016/j.ceb.2013.06.003. Epub 2013 Jul 3. Curr Opin Cell Biol. 2013. PMID: 23830911 Free PMC article. Review.
Cited by
-
Solid stress compression enhances breast cancer cell migration through the upregulation of Interleukin-6.Front Cell Dev Biol. 2025 Apr 30;13:1541953. doi: 10.3389/fcell.2025.1541953. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40371393 Free PMC article.
-
Force-activated zyxin assemblies coordinate actin nucleation and crosslinking to orchestrate stress fiber repair.bioRxiv [Preprint]. 2024 May 18:2024.05.17.594765. doi: 10.1101/2024.05.17.594765. bioRxiv. 2024. Update in: Curr Biol. 2025 Feb 24;35(4):854-870.e9. doi: 10.1016/j.cub.2025.01.042. PMID: 38798419 Free PMC article. Updated. Preprint.
-
Zyxin is important for the stability and function of podocytes, especially during mechanical stretch.Commun Biol. 2024 Apr 11;7(1):446. doi: 10.1038/s42003-024-06125-5. Commun Biol. 2024. PMID: 38605154 Free PMC article.
-
Cellular Traction Force Holds the Potential as a Drug Testing Readout for In Vitro Cancer Metastasis.Cell Mol Bioeng. 2024 Jul 4;17(3):203-217. doi: 10.1007/s12195-024-00811-4. eCollection 2024 Jun. Cell Mol Bioeng. 2024. PMID: 39050509 Free PMC article.
-
Zyxin regulates embryonic stem cell fate by modulating mechanical and biochemical signaling interface.Commun Biol. 2023 Jan 18;6(1):62. doi: 10.1038/s42003-023-04421-0. Commun Biol. 2023. PMID: 36653484 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

