The Making and Breaking of Serine-ADP-Ribosylation in the DNA Damage Response
- PMID: 34869334
- PMCID: PMC8634249
- DOI: 10.3389/fcell.2021.745922
The Making and Breaking of Serine-ADP-Ribosylation in the DNA Damage Response
Abstract
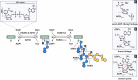
ADP-ribosylation is a widespread posttranslational modification that is of particular therapeutic relevance due to its involvement in DNA repair. In response to DNA damage, PARP1 and 2 are the main enzymes that catalyze ADP-ribosylation at damage sites. Recently, serine was identified as the primary amino acid acceptor of the ADP-ribosyl moiety following DNA damage and appears to act as seed for chain elongation in this context. Serine-ADP-ribosylation strictly depends on HPF1, an auxiliary factor of PARP1/2, which facilitates this modification by completing the PARP1/2 active site. The signal is terminated by initial poly(ADP-ribose) chain degradation, primarily carried out by PARG, while another enzyme, (ADP-ribosyl)hydrolase 3 (ARH3), specifically cleaves the terminal seryl-ADP-ribosyl bond, thus completing the chain degradation initiated by PARG. This review summarizes recent findings in the field of serine-ADP-ribosylation, its mechanisms, possible functions and potential for therapeutic targeting through HPF1 and ARH3 inhibition.
Keywords: ADP-ribosylation; ARH3; DNA damage; PARG; PARP; cancer; neurodegeneration; posttranslational modification (PTM).
Copyright © 2021 Schützenhofer, Rack and Ahel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aberle L., Krüger A., Reber J. M., Lippmann M., Hufnagel M., Schmalz M., et al. (2020). PARP1 catalytic variants reveal branching and chain length-specific functions of poly(ADP-ribose) in cellular physiology and stress response. Nucleic Acids Res. 48 10015–10033. 10.1093/nar/gkaa590 - DOI - PMC - PubMed
-
- Alvarez-Gonzalez R., Jacobson M. K. (1987). Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo1”. Biochemistry 26 3218–3224. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

