Dynamic Expression and Regulatory Network of Circular RNA for Abdominal Preadipocytes Differentiation in Chicken (Gallus gallus)
- PMID: 34869349
- PMCID: PMC8633312
- DOI: 10.3389/fcell.2021.761638
Dynamic Expression and Regulatory Network of Circular RNA for Abdominal Preadipocytes Differentiation in Chicken (Gallus gallus)
Abstract
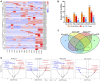
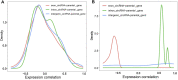
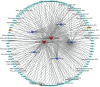
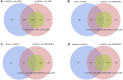
Circular RNA (circRNA), as a novel endogenous biomolecule, has been emergingly demonstrated to play crucial roles in mammalian lipid metabolism and obesity. However, little is known about their genome-wide identification, expression profile, and function in chicken adipogenesis. In present study, the adipogenic differentiation of chicken abdominal preadipocyte was successfully induced, and the regulatory functional circRNAs in chicken adipogenesis were identified from abdominal adipocytes at different differentiation stages using Ribo-Zero RNA-seq. A total of 1,068 circRNA candidates were identified and mostly derived from exons. Of these, 111 differentially expressed circRNAs (DE-circRNAs) were detected, characterized by stage-specific expression, and enriched in several lipid-related pathways, such as Hippo signaling pathway, mTOR signaling pathway. Through weighted gene co-expression network analyses (WGCNA) and K-means clustering analyses, two DE-circRNAs, Z:35565770|35568133 and Z:54674624|54755962, were identified as candidate regulatory circRNAs in chicken adipogenic differentiation. Z:35565770|35568133 might compete splicing with its parental gene, ABHD17B, owing to its strictly negative co-expression. We also constructed competing endogenous RNA (ceRNA) network based on DE-circRNA, DE-miRNA, DE-mRNAs, revealing that Z:54674624|54755962 might function as a ceRNA to regulate chicken adipogenic differentiation through the gga-miR-1635-AHR2/IRF1/MGAT3/ABCA1/AADAC and/or the novel_miR_232-STAT5A axis. Translation activity analysis showed that Z:35565770|35568133 and Z:54674624|54755962 have no protein-coding potential. These findings provide valuable evidence for a better understanding of the specific functions and molecular mechanisms of circRNAs underlying avian adipogenesis.
Keywords: abdominal fat; adipogenic differentiation; chicken; circRNA; competing endogenous RNA.
Copyright © 2021 Tian, Zhang, Zhong, Nie, Ling, Zhang and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008). The microRNA.Org Resource: Targets and Expression. Nucleic Acids Res. 36, D149–D153. 10.1093/nar/gkm995 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

