Systematic review on role of structure based drug design (SBDD) in the identification of anti-viral leads against SARS-Cov-2
- PMID: 34870145
- PMCID: PMC8120892
- DOI: 10.1016/j.crphar.2021.100026
Systematic review on role of structure based drug design (SBDD) in the identification of anti-viral leads against SARS-Cov-2
Abstract
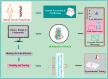
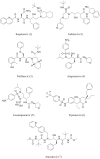
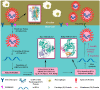
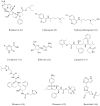
The outbreak of existing public health distress is threatening the entire world with emergence and rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The novel coronavirus disease 2019 (COVID-19) is mild in most people. However, in some elderly people with co-morbid conditions, it may progress to pneumonia, acute respiratory distress syndrome (ARDS) and multi organ dysfunction leading to death. COVID-19 has caused global panic in the healthcare sector and has become one of the biggest threats to the global economy. Drug discovery researchers are expected to contribute rapidly than ever before. The complete genome sequence of coronavirus had been reported barely a month after the identification of first patient. Potential drug targets to combat and treat the coronavirus infection have also been explored. The iterative structure-based drug design (SBDD) approach could significantly contribute towards the discovery of new drug like molecules for the treatment of COVID-19. The existing antivirals and experiences gained from SARS and MERS outbreaks may pave way for identification of potential drug molecules using the approach. SBDD has gained momentum as the essential tool for faster and costeffective lead discovery of antivirals in the past. The discovery of FDA approved human immunodeficiency virus type 1 (HIV-1) inhibitors represent the foremost success of SBDD. This systematic review provides an overview of the novel coronavirus, its pathology of replication, role of structure based drug design, available drug targets and recent advances in in-silico drug discovery for the prevention of COVID-19. SARSCoV- 2 main protease, RNA dependent RNA polymerase (RdRp) and spike (S) protein are the potential targets, which are currently explored for the drug development.
Keywords: Coronavirus 2; Proteases; RNA dependent RNA polymerase; Spike (S) protein; Structure based drug design.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. NGB, SR, GG and AS are thankful to Indian Institute of Technology ( Banaras Hindu University) Varanasi for providing teaching assistantships to them.
Figures

Similar articles
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
-
Development of a Fluorescence-Based, High-Throughput SARS-CoV-2 3CLpro Reporter Assay.J Virol. 2020 Oct 27;94(22):e01265-20. doi: 10.1128/JVI.01265-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32843534 Free PMC article.
-
Approaches and advances in the development of potential therapeutic targets and antiviral agents for the management of SARS-CoV-2 infection.Eur J Pharmacol. 2020 Oct 15;885:173450. doi: 10.1016/j.ejphar.2020.173450. Epub 2020 Jul 31. Eur J Pharmacol. 2020. PMID: 32739174 Free PMC article. Review.
-
Exploring the SARS-Cov-2 Main Protease (Mpro) and RdRp Targets by Updating Current Structure-based Drug Design Utilizing Co-crystals to Combat COVID-19.Curr Drug Targets. 2022;23(8):802-817. doi: 10.2174/1389450122666210906154849. Curr Drug Targets. 2022. PMID: 34488580
Cited by
-
Importance of Computer-aided Drug Design in Modern Pharmaceutical Research.Curr Drug Discov Technol. 2025;22(3):e15701638361318. doi: 10.2174/0115701638361318241230073123. Curr Drug Discov Technol. 2025. PMID: 39810447 Review.
-
Pharmacological therapies and drug development targeting SARS-CoV-2 infection.Cytokine Growth Factor Rev. 2022 Dec;68:13-24. doi: 10.1016/j.cytogfr.2022.10.003. Epub 2022 Oct 13. Cytokine Growth Factor Rev. 2022. PMID: 36266222 Free PMC article. Review.
-
MDFit: automated molecular simulations workflow enables high throughput assessment of ligands-protein dynamics.J Comput Aided Mol Des. 2024 Jul 17;38(1):24. doi: 10.1007/s10822-024-00564-2. J Comput Aided Mol Des. 2024. PMID: 39014286
-
Interference of small compounds and Mg2+ with dsRNA-binding fluorophores compromises the identification of SARS-CoV-2 RdRp inhibitors.Sci Rep. 2024 Nov 15;14(1):28250. doi: 10.1038/s41598-024-78354-x. Sci Rep. 2024. PMID: 39548173 Free PMC article.
-
Discovery of novel IDH1-R132C inhibitors through structure-based virtual screening.Front Pharmacol. 2022 Sep 7;13:982375. doi: 10.3389/fphar.2022.982375. eCollection 2022. Front Pharmacol. 2022. PMID: 36160383 Free PMC article.
References
-
- Agnello S., Brand M., Chellat M.F., Gazzola S., Riedl RJACIE A structural view on medicinal chemistry strategies against drug resistance. 2019;58(11):3300–3345. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

