Potential inhibitors of SARS-CoV-2 (COVID 19) proteases PLpro and Mpro/ 3CLpro: molecular docking and simulation studies of three pertinent medicinal plant natural components
- PMID: 34870149
- PMCID: PMC8178537
- DOI: 10.1016/j.crphar.2021.100038
Potential inhibitors of SARS-CoV-2 (COVID 19) proteases PLpro and Mpro/ 3CLpro: molecular docking and simulation studies of three pertinent medicinal plant natural components
Abstract
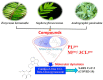
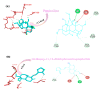
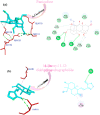
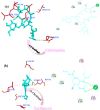
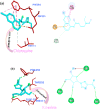
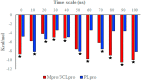
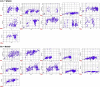
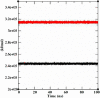
The rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) - coronavirus disease 2019 (COVID-19) has raised a severe global public health issue and creates a pandemic situation. The present work aims to study the molecular -docking and dynamic of three pertinent medicinal plants i.e. Eurycoma harmandiana, Sophora flavescens and Andrographis paniculata phyto-compounds against SARS-COV-2 papain-like protease (PLpro) and main protease (Mpro)/3-chymotrypsin-like protease (3CLpro). The interaction of protein targets and ligands was performed through AutoDock-Vina visualized using PyMOL and BIOVIA-Discovery Studio 2020. Molecular docking with canthin-6-one 9-O-beta-glucopyranoside showed highest binding affinity and less binding energy with both PLpro and Mpro/3CLpro proteases and was subjected to molecular dynamic (MD) simulations for a period of 100ns. Stability of the protein-ligand complexes was evaluated by different analyses. The binding free energy calculated using MM-PBSA and the results showed that the molecule must have stable interactions with the protein binding site. ADMET analysis of the compounds suggested that it is having drug-like properties like high gastrointestinal (GI) absorption, no blood-brain barrier permeability and high lipophilicity. The outcome revealed that canthin-6-one 9-O-beta-glucopyranoside can be used as a potential natural drug against COVID-19 protease.
Keywords: Canthin-6-one 9-O-Beta-glucopyranoside; Molecular dynamics; Natural inhibitor; PLpro-Mpro/3CLpro; SARS-CoV-2 (COVID-19).
© 2021 Published by Elsevier B.V.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures














References
-
- Adeoye A.O., Oso B.J., Olaoye I.F., Tijjani H., Adebayo A.I. Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1765876. - DOI - PMC - PubMed
-
- Andres F. Yepes-Pérez, Oscar Herrera-Calderon, Quintero-Saumeth Jorge. Uncaria tomentosa (cat's claw): a promising herbal medicine against SARS-CoV-2/ACE-2 junction and SARS-CoV-2 spike protein based on molecular modeling. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1837676. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

