Rationally designed mariner vectors for functional genomic analysis of Actinobacillus pleuropneumoniae and other Pasteurellaceae species by transposon-directed insertion-site sequencing (TraDIS)
- PMID: 34870287
- PMCID: PMC8616859
- DOI: 10.1186/s44149-021-00026-4
Rationally designed mariner vectors for functional genomic analysis of Actinobacillus pleuropneumoniae and other Pasteurellaceae species by transposon-directed insertion-site sequencing (TraDIS)
Abstract
Comprehensive identification of conditionally essential genes requires efficient tools for generating high-density transposon libraries that, ideally, can be analysed using next-generation sequencing methods such as Transposon Directed Insertion-site Sequencing (TraDIS). The Himar1 (mariner) transposon is ideal for generating near-saturating mutant libraries, especially in AT-rich chromosomes, as the requirement for integration is a TA dinucleotide, and this transposon has been used for mutagenesis of a wide variety of bacteria. However, plasmids for mariner delivery do not necessarily work well in all bacteria. In particular, there are limited tools for functional genomic analysis of Pasteurellaceae species of major veterinary importance, such as swine and cattle pathogens, Actinobacillus pleuropneumoniae and Pasteurella multocida, respectively. Here, we developed plasmids, pTsodCPC9 and pTlacPC9 (differing only in the promoter driving expression of the transposase gene), that allow delivery of mariner into both these pathogens, but which should also be applicable to a wider range of bacteria. Using the pTlacPC9 vector, we have generated, for the first time, saturating mariner mutant libraries in both A. pleuropneumoniae and P. multocida that showed a near random distribution of insertions around the respective chromosomes as detected by TraDIS. A preliminary screen of 5000 mutants each identified 8 and 14 genes, respectively, that are required for growth under anaerobic conditions. Future high-throughput screening of the generated libraries will facilitate identification of mutants required for growth under different conditions, including in vivo, highlighting key virulence factors and pathways that can be exploited for development of novel therapeutics and vaccines.
Keywords: Actinobacillus pleuropneumoniae; Mariner; Pasteurella multocida; Pasteurellaceae; TraDIS; Transposon.
© The Author(s) 2021.
Conflict of interest statement
Competing interestsAuthor Paul R. Langford was not involved in the journal’s review or decisions related to this manuscript. The authors declare no competing interests.
Figures
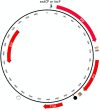
 ), and the origin of plasmid replication (colE1) as a hollow lollipop (
), and the origin of plasmid replication (colE1) as a hollow lollipop ( ). Coloured blocks flanking the cat gene indicate the locations of Himar1 inverted repeats (thick orange), and paired copies of DNA uptake sequences for Neisseria spp. (thin green), H. influenzae (thin pink) and A. pleuropneumoniae (thin blue). Arrowhead upstream of the C9 tnp gene indicates promoter sequences for either A. pleuropneumoniae sodC or E. coli lac genes, depending on the vector (pTsodCPC9 and pTlacPC9, respectively)
). Coloured blocks flanking the cat gene indicate the locations of Himar1 inverted repeats (thick orange), and paired copies of DNA uptake sequences for Neisseria spp. (thin green), H. influenzae (thin pink) and A. pleuropneumoniae (thin blue). Arrowhead upstream of the C9 tnp gene indicates promoter sequences for either A. pleuropneumoniae sodC or E. coli lac genes, depending on the vector (pTsodCPC9 and pTlacPC9, respectively)
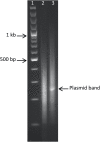
References
-
- Baltes N, Buettner FFR, Gerlach G-F. Selective capture of transcribed sequences (SCOTS) of Actinobacillus pleuropneumoniae in the chronic stage of disease reveals an HlyX-regulated autotransporter protein. Veterinary Microbiology. 2007;123(1–3):110–121. doi: 10.1016/j.vetmic.2007.03.026. - DOI - PubMed
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
-
- Bossé JT, Chaudhuri RR, Li Y, Leanse LG, Fernandez Crespo R, Coupland P, Holden MTG, Bazzolli DM, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, Langford PR, on behalf of the BRaDP1T consortium Complete genome sequence of MIDG2331, a genetically tractable serovar 8 clinical isolate of Actinobacillus pleuropneumoniae. Genome Announcements. 2016;4(1):e01667–e01615. doi: 10.1128/genomeA.01667-15. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
