iProX in 2021: connecting proteomics data sharing with big data
- PMID: 34871441
- PMCID: PMC8728291
- DOI: 10.1093/nar/gkab1081
iProX in 2021: connecting proteomics data sharing with big data
Abstract
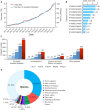
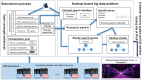
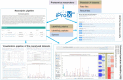
The rapid development of proteomics studies has resulted in large volumes of experimental data. The emergence of big data platform provides the opportunity to handle these large amounts of data. The integrated proteome resource, iProX (https://www.iprox.cn), which was initiated in 2017, has been greatly improved with an up-to-date big data platform implemented in 2021. Here, we describe the main iProX developments since its first publication in Nucleic Acids Research in 2019. First, a hyper-converged architecture with high scalability supports the submission process. A hadoop cluster can store large amounts of proteomics datasets, and a distributed, RESTful-styled Elastic Search engine can query millions of records within one second. Also, several new features, including the Universal Spectrum Identifier (USI) mechanism proposed by ProteomeXchange, RESTful Web Service API, and a high-efficiency reanalysis pipeline, have been added to iProX for better open data sharing. By the end of August 2021, 1526 datasets had been submitted to iProX, reaching a total data volume of 92.42TB. With the implementation of the big data platform, iProX can support PB-level data storage, hundreds of billions of spectra records, and second-level latency service capabilities that meet the requirements of the fast growing field of proteomics.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Marx V. Biology: the big challenges of big data. Nature. 2013; 498:255–260. - PubMed
-
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M.et al. .. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

