Plasma Membrane Anchoring and Gag:Gag Multimerization on Viral RNA Are Critical Properties of HIV-1 Gag Required To Mediate Efficient Genome Packaging
- PMID: 34872357
- PMCID: PMC8649766
- DOI: 10.1128/mbio.03254-21
Plasma Membrane Anchoring and Gag:Gag Multimerization on Viral RNA Are Critical Properties of HIV-1 Gag Required To Mediate Efficient Genome Packaging
Abstract
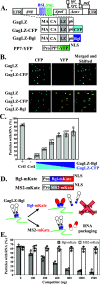
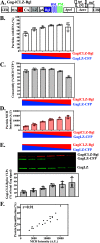
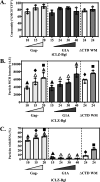
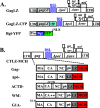
Human immunodeficiency virus type 1 (HIV-1) Gag selects and packages the HIV RNA genome during virus assembly. However, HIV-1 RNA constitutes only a small fraction of the cellular RNA. Although Gag exhibits a slight preference to viral RNA, most of the cytoplasmic Gag proteins are associated with cellular RNAs. Thus, it is not understood how HIV-1 achieves highly efficient genome packaging. We hypothesize that besides RNA binding, other properties of Gag are important for genome packaging. Many Gag mutants have assembly defects that preclude analysis of their effects on genome packaging. To bypass this challenge, we established complementation systems that separate the particle-assembling and RNA-binding functions of Gag: we used a set of Gag proteins to drive particle assembly and an RNA-binding Gag to package HIV-1 RNA. We have developed two types of RNA-binding Gag in which packaging is mediated by the authentic nucleocapsid (NC) domain or by a nonviral RNA-binding domain. We found that in both cases, mutations that affect the multimerization or plasma membrane anchoring properties of Gag reduce or abolish RNA packaging. These mutant Gag can coassemble into particles but cannot package the RNA genome efficiently. Our findings indicate that HIV-1 RNA packaging occurs at the plasma membrane and RNA-binding Gag needs to multimerize on RNA to encapsidate the viral genome. IMPORTANCE To generate infectious virions, HIV-1 must package its full-length RNA as the genome during particle assembly. HIV-1 Gag:RNA interactions mediate genome packaging, but the mechanism remains unclear. Only a minor portion of the cellular RNA is HIV-1 RNA, and most of the RNAs associated with cytoplasmic Gag are cellular RNAs. However, >94% of the HIV-1 virions contain viral RNA genome. We posited that, besides RNA binding, other properties of Gag contribute to genome packaging. Using two complementation systems, we examined features of Gag that are important for genome packaging. We found that the capacities for Gag to multimerize and to anchor at the plasma membrane are critical for genome packaging. Our results revealed that Gag needs to multimerize on viral RNA at the plasma membrane in order to package RNA genome.
Keywords: Gag; Gag:RNA interactions; RNA; genome packaging; human immunodeficiency virus; membrane targeting; multimerization.
Figures


Similar articles
-
Unpaired Guanosines in the 5' Untranslated Region of HIV-1 RNA Act Synergistically To Mediate Genome Packaging.J Virol. 2020 Oct 14;94(21):e00439-20. doi: 10.1128/JVI.00439-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796062 Free PMC article.
-
Interactions between HIV-1 Gag and Viral RNA Genome Enhance Virion Assembly.J Virol. 2017 Jul 27;91(16):e02319-16. doi: 10.1128/JVI.02319-16. Print 2017 Aug 15. J Virol. 2017. PMID: 28539452 Free PMC article.
-
Subcellular Localization of HIV-1 gag-pol mRNAs Regulates Sites of Virion Assembly.J Virol. 2017 Feb 28;91(6):e02315-16. doi: 10.1128/JVI.02315-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28053097 Free PMC article.
-
Rendezvous at Plasma Membrane: Cellular Lipids and tRNA Set up Sites of HIV-1 Particle Assembly and Incorporation of Host Transmembrane Proteins.Viruses. 2020 Jul 31;12(8):842. doi: 10.3390/v12080842. Viruses. 2020. PMID: 32752131 Free PMC article. Review.
-
HIV-1 RNA genome packaging: it's G-rated.mBio. 2024 Apr 10;15(4):e0086123. doi: 10.1128/mbio.00861-23. Epub 2024 Feb 27. mBio. 2024. PMID: 38411060 Free PMC article. Review.
Cited by
-
Single-Virion Analysis: A Method to Visualize HIV-1 Particle Content Using Fluorescence Microscopy.Methods Mol Biol. 2024;2807:77-91. doi: 10.1007/978-1-0716-3862-0_6. Methods Mol Biol. 2024. PMID: 38743222
-
Specific Interaction of DARPin with HIV-1 CANTD Disturbs the Distribution of Gag, RNA Packaging, and Tetraspanin Remodelling in the Membrane.Viruses. 2022 Apr 15;14(4):824. doi: 10.3390/v14040824. Viruses. 2022. PMID: 35458554 Free PMC article.
-
Conformational transitions of the HIV-1 Gag polyprotein upon multimerization and gRNA binding.Biophys J. 2024 Jan 2;123(1):42-56. doi: 10.1016/j.bpj.2023.11.017. Epub 2023 Nov 18. Biophys J. 2024. PMID: 37978800 Free PMC article.
-
Relationship between HIV-1 Gag Multimerization and Membrane Binding.Viruses. 2022 Mar 16;14(3):622. doi: 10.3390/v14030622. Viruses. 2022. PMID: 35337029 Free PMC article. Review.
-
Human Retrovirus Genomic RNA Packaging.Viruses. 2022 May 19;14(5):1094. doi: 10.3390/v14051094. Viruses. 2022. PMID: 35632835 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

