Whole-genome resequencing of three Coilia nasus population reveals genetic variations in genes related to immune, vision, migration, and osmoregulation
- PMID: 34872488
- PMCID: PMC8647404
- DOI: 10.1186/s12864-021-08182-0
Whole-genome resequencing of three Coilia nasus population reveals genetic variations in genes related to immune, vision, migration, and osmoregulation
Abstract
Background: Coilia nasus is an important anadromous fish, widely distributed in China, Japan, and Korea. Based on morphological and ecological researches of C. nasus, two ecotypes were identified. One is the anadromous population (AP). The sexually mature fish run thousands of kilometers from marine to river for spawning. Another one is the resident population which cannot migrate. Based on their different habitats, they were classified into landlocked population (LP) and sea population (SP) which were resident in the freshwater lake and marine during the entire lifetime, respectively. However, they have never been systematically studied. Moreover, C. nasus is declining sharply due to overfishing and pollution recently. Therefore, further understandings of C. nasus populations are needed for germplasm protection.
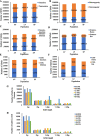
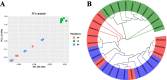
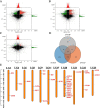
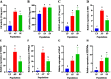
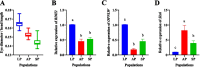
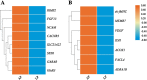
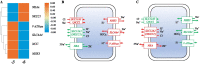
Results: Whole-genome resequencing of AP, LP, and SP were performed to enrich the understanding of different populations of C. nasus. At the genome level, 3,176,204, 3,307,069, and 3,207,906 single nucleotide polymorphisms (SNPs) and 1,892,068, 2,002,912, and 1,922,168 insertion/deletion polymorphisms (InDels) were generated in AP, LP, and SP, respectively. Selective sweeping analysis showed that 1022 genes were selected in AP vs LP; 983 genes were selected in LP vs SP; 116 genes were selected in AP vs SP. Among them, selected genes related to immune, vision, migration, and osmoregulation were identified. Furthermore, their expression profiles were detected by quantitative real-time PCR. Expression levels of selected genes related to immune, and vision in LP were significantly lower than AP and SP. Selected genes related to migration in AP were expressed significantly more highly than LP. Expression levels of selected genes related to osmoregulation were also detected. The expression of NKAα and NKCC1 in LP were significantly lower than SP, while expression of NCC, SLC4A4, NHE3, and V-ATPase in LP was significantly higher than SP.
Conclusions: Combined to life history of C. nasus populations, our results revealed that the molecular mechanisms of their differences of immune, vision, migration, and osmoregulation. Our findings will provide a further understanding of different populations of C. nasus and will be beneficial for wild C. nasus protection.
Keywords: Coilia nasus populations; Genetic variation; Genome resequencing; Migration; Selective sweeping.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Loss of Coilia nasus habitats in Chinese freshwater lakes: An otolith microchemistry assessment.Heliyon. 2020 Aug 1;6(8):e04571. doi: 10.1016/j.heliyon.2020.e04571. eCollection 2020 Aug. Heliyon. 2020. PMID: 32775746 Free PMC article.
-
Population genetic structure of the acanthocephalan Acanthosentis cheni in anadromous, freshwater, and landlocked stocks of its fish host, Coilia nasus.J Parasitol. 2014 Apr;100(2):193-7. doi: 10.1645/12-144.1. Epub 2013 Nov 13. J Parasitol. 2014. PMID: 24224788
-
Transcriptome analysis of the brain provides insights into the regulatory mechanism for Coilia nasus migration.BMC Genomics. 2020 Jun 18;21(1):410. doi: 10.1186/s12864-020-06816-3. BMC Genomics. 2020. PMID: 32552858 Free PMC article.
-
Identification of a uniquely expanded V1R (ORA) gene family in the Japanese grenadier anchovy (Coilia nasus).Mar Biol. 2016;163:126. doi: 10.1007/s00227-016-2896-9. Epub 2016 May 2. Mar Biol. 2016. PMID: 27340293 Free PMC article.
-
Regulation of signal transduction in Coilia nasus during migration.Genomics. 2020 Jan;112(1):55-64. doi: 10.1016/j.ygeno.2019.07.021. Epub 2019 Aug 9. Genomics. 2020. PMID: 31404627
Cited by
-
Genome-wide identification of the NHE gene family in Coilia nasus and its response to salinity challenge and ammonia stress.BMC Genomics. 2022 Jul 20;23(1):526. doi: 10.1186/s12864-022-08761-9. BMC Genomics. 2022. PMID: 35858854 Free PMC article.
-
Whole-genome re-sequencing provides key genomic insights in farmed Arctic charr (Salvelinus alpinus) populations of anadromous and landlocked origin from Scandinavia.Evol Appl. 2023 Feb 27;16(4):797-813. doi: 10.1111/eva.13537. eCollection 2023 Apr. Evol Appl. 2023. PMID: 37124091 Free PMC article.
References
-
- Jiang T, Yang J, Liu H, Shen X, qiang. Life history of Coilia nasus from the Yellow Sea inferred from otolith Sr:Ca ratios. Environ Biol Fish. 2012;95:503–508. doi: 10.1007/s10641-012-0066-6. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

