Distinct roles of hnRNPH1 low-complexity domains in splicing and transcription
- PMID: 34873036
- PMCID: PMC8685725
- DOI: 10.1073/pnas.2109668118
Distinct roles of hnRNPH1 low-complexity domains in splicing and transcription
Abstract
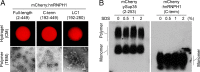
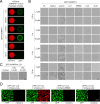
Heterogeneous nuclear ribonucleoproteins (hnRNPs) represent a large family of RNA-binding proteins that control key events in RNA biogenesis under both normal and diseased cellular conditions. The low-complexity (LC) domain of hnRNPs can become liquid-like droplets or reversible amyloid-like polymers by phase separation. Yet, whether phase separation of the LC domains contributes to physiological functions of hnRNPs remains unclear. hnRNPH1 contains two LC domains, LC1 and LC2. Here, we show that reversible phase separation of the LC1 domain is critical for both interaction with different kinds of RNA-binding proteins and control of the alternative-splicing activity of hnRNPH1. Interestingly, although not required for phase separation, the LC2 domain contributes to the robust transcriptional activation of hnRNPH1 when fused to the DNA-binding domain, as found recently in acute lymphoblastic leukemia. Our data suggest that the ability of the LC1 domain to phase-separate into reversible polymers or liquid-like droplets is essential for function of hnRNPH1 as an alternative RNA-splicing regulator, whereas the LC2 domain may contribute to the aberrant transcriptional activity responsible for cancer transformation.
Keywords: hnRNPH1; low-complexity domain; phase separation; splicing; transcription.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Kato M., McKnight S. L., A solid-state conceptualization of information transfer from gene to message to protein. Annu. Rev. Biochem. 87, 351–390 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

