Class-specific interactions between Sis1 J-domain protein and Hsp70 chaperone potentiate disaggregation of misfolded proteins
- PMID: 34873058
- PMCID: PMC8670446
- DOI: 10.1073/pnas.2108163118
Class-specific interactions between Sis1 J-domain protein and Hsp70 chaperone potentiate disaggregation of misfolded proteins
Abstract
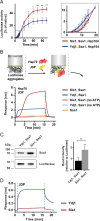
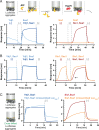
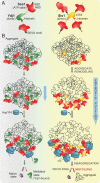
Protein homeostasis is constantly being challenged with protein misfolding that leads to aggregation. Hsp70 is one of the versatile chaperones that interact with misfolded proteins and actively support their folding. Multifunctional Hsp70s are harnessed to specific roles by J-domain proteins (JDPs, also known as Hsp40s). Interaction with the J-domain of these cochaperones stimulates ATP hydrolysis in Hsp70, which stabilizes substrate binding. In eukaryotes, two classes of JDPs, Class A and Class B, engage Hsp70 in the reactivation of aggregated proteins. In most species, excluding metazoans, protein recovery also relies on an Hsp100 disaggregase. Although intensely studied, many mechanistic details of how the two JDP classes regulate protein disaggregation are still unknown. Here, we explore functional differences between the yeast Class A (Ydj1) and Class B (Sis1) JDPs at the individual stages of protein disaggregation. With real-time biochemical tools, we show that Ydj1 alone is superior to Sis1 in aggregate binding, yet it is Sis1 that recruits more Ssa1 molecules to the substrate. This advantage of Sis1 depends on its ability to bind to the EEVD motif of Hsp70, a quality specific to most of Class B JDPs. This second interaction also conditions the Hsp70-induced aggregate modification that boosts its subsequent dissolution by the Hsp104 disaggregase. Our results suggest that the Sis1-mediated chaperone assembly at the aggregate surface potentiates the entropic pulling, driven polypeptide disentanglement, while Ydj1 binding favors the refolding of the solubilized proteins. Such subspecialization of the JDPs across protein reactivation improves the robustness and efficiency of the disaggregation machinery.
Keywords: Hsp40; chaperones; protein aggregation.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
NMR Studies on the Structure of Yeast Sis1 and the Dynamics of Its Interaction with Ssa1-EEVD.Molecules. 2024 Dec 24;30(1):11. doi: 10.3390/molecules30010011. Molecules. 2024. PMID: 39795068 Free PMC article.
-
Role of J-domain Proteins in Yeast Physiology and Protein Quality Control.J Mol Biol. 2024 Jul 15;436(14):168484. doi: 10.1016/j.jmb.2024.168484. Epub 2024 Feb 7. J Mol Biol. 2024. PMID: 38331212 Review.
-
Roles of intramolecular and intermolecular interactions in functional regulation of the Hsp70 J-protein co-chaperone Sis1.J Mol Biol. 2015 Apr 10;427(7):1632-43. doi: 10.1016/j.jmb.2015.02.007. Epub 2015 Feb 14. J Mol Biol. 2015. PMID: 25687964 Free PMC article.
-
Early steps of protein disaggregation by Hsp70 chaperone and class B J-domain proteins are shaped by Hsp110.Elife. 2024 Oct 15;13:RP94795. doi: 10.7554/eLife.94795. Elife. 2024. PMID: 39404743 Free PMC article.
-
Three J-proteins impact Hsp104-mediated variant-specific prion elimination: a new critical role for a low-complexity domain.Curr Genet. 2020 Feb;66(1):51-58. doi: 10.1007/s00294-019-01006-5. Epub 2019 Jun 22. Curr Genet. 2020. PMID: 31230108 Free PMC article. Review.
Cited by
-
Comparative genomic analysis reveals expansion of the DnaJ gene family in Lagerstroemia indica and its members response to salt stress.Genetica. 2024 Jun;152(2-3):101-117. doi: 10.1007/s10709-024-00208-1. Epub 2024 May 10. Genetica. 2024. PMID: 38724749
-
Design principles to tailor Hsp104 therapeutics.bioRxiv [Preprint]. 2024 Apr 28:2024.04.26.591398. doi: 10.1101/2024.04.26.591398. bioRxiv. 2024. Update in: Cell Rep. 2024 Dec 24;43(12):115005. doi: 10.1016/j.celrep.2024.115005. PMID: 38712168 Free PMC article. Updated. Preprint.
-
J-domain proteins: From molecular mechanisms to diseases.Cell Stress Chaperones. 2024 Feb;29(1):21-33. doi: 10.1016/j.cstres.2023.12.002. Epub 2023 Dec 23. Cell Stress Chaperones. 2024. PMID: 38320449 Free PMC article.
-
Advances in the study of HSP70 inhibitors to enhance the sensitivity of tumor cells to radiotherapy.Front Cell Dev Biol. 2022 Aug 10;10:942828. doi: 10.3389/fcell.2022.942828. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36036010 Free PMC article. Review.
-
Distribution and solvent exposure of Hsp70 chaperone binding sites across the Escherichia coli proteome.Proteins. 2023 May;91(5):665-678. doi: 10.1002/prot.26456. Epub 2023 Jan 1. Proteins. 2023. PMID: 36539330 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

