Medium levels of transcription and replication related chromosomal instability are associated with poor clinical outcome
- PMID: 34873180
- PMCID: PMC8648741
- DOI: 10.1038/s41598-021-02787-x
Medium levels of transcription and replication related chromosomal instability are associated with poor clinical outcome
Abstract
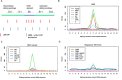
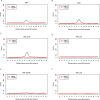
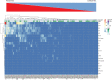
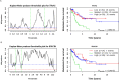
Genomic instability (GI) influences treatment efficacy and resistance, and an accurate measure of it is lacking. Current measures of GI are based on counts of specific structural variation (SV) and mutational signatures. Here, we present a holistic approach to measuring GI based on the quantification of the steady-state equilibrium between DNA damage and repair as assessed by the residual breakpoints (BP) remaining after repair, irrespective of SV type. We use the notion of Hscore, a BP "hotspotness" magnitude scale, to measure the propensity of genomic structural or functional DNA elements to break more than expected by chance. We then derived new measures of transcription- and replication-associated GI that we call iTRAC (transcription-associated chromosomal instability index) and iRACIN (replication-associated chromosomal instability index). We show that iTRAC and iRACIN are predictive of metastatic relapse in Leiomyosarcoma (LMS) and that they may be combined to form a new classifier called MAGIC (mixed transcription- and replication-associated genomic instability classifier). MAGIC outperforms the gold standards FNCLCC and CINSARC in stratifying metastatic risk in LMS. Furthermore, iTRAC stratifies chemotherapeutic response in LMS. We finally show that this approach is applicable to other cancers.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest. Request for Grant of a European patent has been submitted under the application number: EP20306558.6. the applicants are: Institut National de la Santé et de la Recherche Médicale, UNIVERSITÉ PAUL SABATIER TOULOUSE III, Benhaddou Ataaillah, and Institut Claudius Regaud. The inventors are not yet designated. The Extended european search report is closed. The application covers the aspects of the manuscript relating to hotspots detection, iTRAC and iRACIN indexes, and a method for stratification of clinical outcome based on those indexes.
Figures




References
-
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. - PubMed
-
- Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat. Rev. Cancer. 2015;15:276–289. - PubMed
-
- Macheret M, Halazonetis TD. DNA Replication stress as a hallmark of cancer. Annu. Rev. Pathol. Mech. Dis. 2015;10:425–448. - PubMed
-
- Técher H, Koundrioukoff S, Nicolas A, Debatisse M. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat. Rev. Genet. 2017;18:535–550. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

