Peptides derived from gp43, the most antigenic protein from Paracoccidioides brasiliensis, form amyloid fibrils in vitro: implications for vaccine development
- PMID: 34873233
- PMCID: PMC8648789
- DOI: 10.1038/s41598-021-02898-5
Peptides derived from gp43, the most antigenic protein from Paracoccidioides brasiliensis, form amyloid fibrils in vitro: implications for vaccine development
Abstract
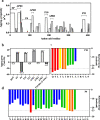
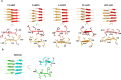
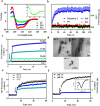
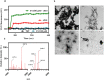
Fungal infection is an important health problem in Latin America, and in Brazil in particular. Paracoccidioides (mainly P. brasiliensis and P. lutzii) is responsible for paracoccidioidomycosis, a disease that affects mainly the lungs. The glycoprotein gp43 is involved in fungi adhesion to epithelial cells, which makes this protein an interesting target of study. A specific stretch of 15 amino acids that spans the region 181-195 (named P10) of gp43 is an important epitope of gp43 that is being envisioned as a vaccine candidate. Here we show that synthetic P10 forms typical amyloid aggregates in solution in very short times, a property that could hamper vaccine development. Seeds obtained by fragmentation of P10 fibrils were able to induce the aggregation of P4, but not P23, two other peptides derived from gp43. In silico analysis revealed several regions within the P10 sequence that can form amyloid with steric zipper architecture. Besides, in-silico proteolysis studies with gp43 revealed that aggregation-prone, P10-like peptides could be generated by several proteases, which suggests that P10 could be formed under physiological conditions. Considering our data in the context of a potential vaccine development, we redesigned the sequence of P10, maintaining the antigenic region (HTLAIR), but drastically reducing its aggregation propensity.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Cordova L. A., Torres J. Paracoccidioidomycosis in StatPearls. (StatPearls Publishing, 2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

