Comparative assessment of favipiravir and remdesivir against human coronavirus NL63 in molecular docking and cell culture models
- PMID: 34873274
- PMCID: PMC8648821
- DOI: 10.1038/s41598-021-02972-y
Comparative assessment of favipiravir and remdesivir against human coronavirus NL63 in molecular docking and cell culture models
Abstract
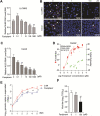
Human coronavirus NL63 (HCoV-NL63) mainly affects young children and immunocompromised patients, causing morbidity and mortality in a subset of patients. Since no specific treatment is available, this study aims to explore the anti-SARS-CoV-2 agents including favipiravir and remdesivir for treating HCoV-NL63 infection. We first successfully modelled the 3D structure of HCoV-NL63 RNA-dependent RNA polymerase (RdRp) based on the experimentally solved SARS-CoV-2 RdRp structure. Molecular docking indicated that favipiravir has similar binding affinities to SARS-CoV-2 and HCoV-NL63 RdRp with LibDock scores of 75 and 74, respectively. The LibDock scores of remdesivir to SARS-CoV-2 and HCoV-NL63 were 135 and 151, suggesting that remdesivir may have a higher affinity to HCoV-NL63 compared to SARS-CoV-2 RdRp. In cell culture models infected with HCoV-NL63, both favipiravir and remdesivir significantly inhibited viral replication and production of infectious viruses. Overall, remdesivir compared to favipiravir is more potent in inhibiting HCoV-NL63 in cell culture. Importantly, there is no evidence of resistance development upon long-term exposure to remdesivir. Furthermore, combining favipiravir or remdesivir with the clinically used antiviral cytokine interferon-alpha resulted in synergistic effects. These findings provided a proof-of-concept that anti-SARS-CoV-2 drugs, in particular remdesivir, have the potential to be repurposed for treating HCoV-NL63 infection.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- da Veiga ABG, Martins LG, Riediger I, Mazetto A, Debur MD, Gregianini TS. More than just a common cold: Endemic coronaviruses OC43, HKU1, NL63, and 229E associated with severe acute respiratory infection and fatality cases among healthy adults. J. Med. Virol. 2020;93:1002–1007. doi: 10.1002/jmv.26362. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

