T cells targeted to TdT kill leukemic lymphoblasts while sparing normal lymphocytes
- PMID: 34873326
- PMCID: PMC9005346
- DOI: 10.1038/s41587-021-01089-x
T cells targeted to TdT kill leukemic lymphoblasts while sparing normal lymphocytes
Abstract
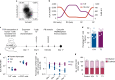
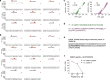
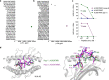
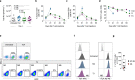
Unlike chimeric antigen receptors, T-cell receptors (TCRs) can recognize intracellular targets presented on human leukocyte antigen (HLA) molecules. Here we demonstrate that T cells expressing TCRs specific for peptides from the intracellular lymphoid-specific enzyme terminal deoxynucleotidyl transferase (TdT), presented in the context of HLA-A*02:01, specifically eliminate primary acute lymphoblastic leukemia (ALL) cells of T- and B-cell origin in vitro and in three mouse models of disseminated B-ALL. By contrast, the treatment spares normal peripheral T- and B-cell repertoires and normal myeloid cells in vitro, and in vivo in humanized mice. TdT is an attractive cancer target as it is highly and homogeneously expressed in 80-94% of B- and T-ALLs, but only transiently expressed during normal lymphoid differentiation, limiting on-target toxicity of TdT-specific T cells. TCR-modified T cells targeting TdT may be a promising immunotherapy for B-ALL and T-ALL that preserves normal lymphocytes.
© 2021. The Author(s).
Conflict of interest statement
J.O. has a research collaboration with Kite Pharma/Gilead, Inc., and a patent application by the institutional technology transfer office Inven2 was filed (patent application number 2113437.4) that covers TCR receptor sequences (inventors J.O. and M.A.). T.N.S. is a consultant for Adaptive Biotechnologies, AIMM Therapeutics, Allogene Therapeutics, Merus, Neogene Therapeutics, Neon Therapeutics and Scenic Biotech; is a recipient of grant/research support from MSD, Bristol-Myers Squibb and Merck KGaA; is a stockholder in AIMM Therapeutics, Allogene Therapeutics, Merus, Neogene Therapeutics and Neon Therapeutics; and is venture partner at Third Rock Ventures. G.E.T. has received research support from Mundipharma and lecture honoraria from Mundipharma, Abbvie, Janssen-Cilag, Alexion Pharmaceuticals and Roche Norway. The remaining authors declare no competing interests.
Figures












References
-
- Grupp SA, et al. Updated analysis of the efficacy and safety of tisagenlecleucel in pediatric and young adult patients with relapsed/refractory (r/r) acute lymphoblastic leukemia. Blood. 2018;132:895–895. doi: 10.1182/blood-2018-99-112599. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

