Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology
- PMID: 34873335
- PMCID: PMC8648564
- DOI: 10.1038/s41588-021-00973-1
Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology
Erratum in
-
Author Correction: Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology.Nat Genet. 2022 Mar;54(3):361. doi: 10.1038/s41588-022-01020-3. Nat Genet. 2022. PMID: 35102318 Free PMC article. No abstract available.
Abstract
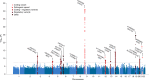
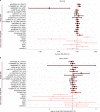
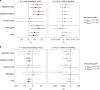
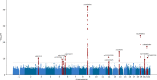
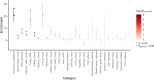
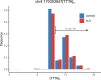
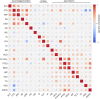
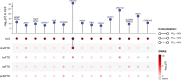
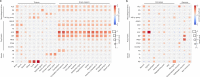
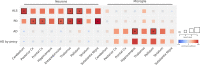
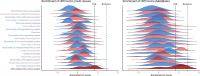
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease with a lifetime risk of one in 350 people and an unmet need for disease-modifying therapies. We conducted a cross-ancestry genome-wide association study (GWAS) including 29,612 patients with ALS and 122,656 controls, which identified 15 risk loci. When combined with 8,953 individuals with whole-genome sequencing (6,538 patients, 2,415 controls) and a large cortex-derived expression quantitative trait locus (eQTL) dataset (MetaBrain), analyses revealed locus-specific genetic architectures in which we prioritized genes either through rare variants, short tandem repeats or regulatory effects. ALS-associated risk loci were shared with multiple traits within the neurodegenerative spectrum but with distinct enrichment patterns across brain regions and cell types. Of the environmental and lifestyle risk factors obtained from the literature, Mendelian randomization analyses indicated a causal role for high cholesterol levels. The combination of all ALS-associated signals reveals a role for perturbations in vesicle-mediated transport and autophagy and provides evidence for cell-autonomous disease initiation in glutamatergic neurons.
© 2021. The Author(s).
Conflict of interest statement
J.H.V. has sponsored research agreements with Biogen. L.H.v.d.B. receives personal fees from Cytokinetics outside of the submitted work. A.A.-C. has served on scientific advisory boards for Mitsubishi Tanabe Pharma, Orion Pharma, Biogen, Lilly, GSK, Apellis, Amylyx and Wave Therapeutics. A. Chiò. serves on scientific advisory boards for Mitsubishi Tanabe, Roche, Biogen, Denali and Cytokinetics. J.E.L. is a member of the scientific advisory board for Cerevel Therapeutics, a consultant for ACI Clinical LLC sponsored by Biogen, Inc. or Ionis Pharmaceuticals, Inc. J.E.L. is also a consultant for Perkins Coie LLP and may provide expert testimony. The remaining authors declare no competing interests related to this work.
Figures




Comment in
-
Translating amyotrophic lateral sclerosis genes into drug development leads.Nat Genet. 2021 Dec;53(12):1624-1626. doi: 10.1038/s41588-021-00981-1. Nat Genet. 2021. PMID: 34873336 No abstract available.
-
Amyotrophic lateral sclerosis - insight into susceptibility.Nat Rev Neurol. 2022 Apr;18(4):189-190. doi: 10.1038/s41582-022-00629-6. Nat Rev Neurol. 2022. PMID: 35197579 No abstract available.
References
-
- van Es MA, et al. Amyotrophic lateral sclerosis. Lancet. 2017;390:2084–2098. - PubMed
-
- Al-Chalabi A, van den Berg LH, Veldink JH. Gene discovery in amyotrophic lateral sclerosis: implications for clinical management. Nat. Rev. Neurol. 2017;13:96–104. - PubMed
-
- Byrne S, Elamin M, Bede P, Hardiman O. Absence of consensus in diagnostic criteria for familial neurodegenerative diseases. J. Neurol. Neurosurg. Psychiatry. 2012;83:365–367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/L501542/1/MRC_/Medical Research Council/United Kingdom
- R56 NS073873/NS/NINDS NIH HHS/United States
- MR/L501529/1/MRC_/Medical Research Council/United Kingdom
- G0701075/MRC_/Medical Research Council/United Kingdom
- G-1307/PUK_/Parkinson's UK/United Kingdom
- MC_UU_00011/1/MRC_/Medical Research Council/United Kingdom
- G0500289/MRC_/Medical Research Council/United Kingdom
- SMITH/APR16/847-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- MR/J004758/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_17115/MRC_/Medical Research Council/United Kingdom
- MC_UU_00011/4/MRC_/Medical Research Council/United Kingdom
- MR/R024804/1/MRC_/Medical Research Council/United Kingdom
- G0901254/MRC_/Medical Research Council/United Kingdom
- G0900635/MRC_/Medical Research Council/United Kingdom
- SHAW/APR15/970-797/MNDA_/Motor Neurone Disease Association/United Kingdom
- ALCHALABI-DOBSON/APR14/829-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- MC_G1000733/MRC_/Medical Research Council/United Kingdom
- MR/K01417X/1/MRC_/Medical Research Council/United Kingdom
- G-0907/PUK_/Parkinson's UK/United Kingdom
- G0600974/MRC_/Medical Research Council/United Kingdom
- G1001253/MRC_/Medical Research Council/United Kingdom
- R01 NS073873/NS/NINDS NIH HHS/United States
- MR/L021803/1/MRC_/Medical Research Council/United Kingdom
- G1100695/MRC_/Medical Research Council/United Kingdom
- ALCHALABI-TALBOT/APR14/926-794/MNDA_/Motor Neurone Disease Association/United Kingdom
- G0900688/MRC_/Medical Research Council/United Kingdom
- MR/L023784/2/MRC_/Medical Research Council/United Kingdom
- MRF-060-0003-RG-SMITH/MRF_/MRF_/United Kingdom
- SHAW/NOV14/985-797/MNDA_/Motor Neurone Disease Association/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

