This is a preprint.
Omicron (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance
- PMID: 34873578
- PMCID: PMC8647651
Omicron (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance
Update in
-
Omicron Variant (B.1.1.529): Infectivity, Vaccine Breakthrough, and Antibody Resistance.J Chem Inf Model. 2022 Jan 24;62(2):412-422. doi: 10.1021/acs.jcim.1c01451. Epub 2022 Jan 6. J Chem Inf Model. 2022. PMID: 34989238 Free PMC article.
Abstract
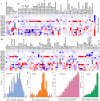
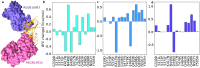
The latest severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant Omicron (B.1.1.529) has ushered panic responses around the world due to its contagious and vaccine escape mutations. The essential infectivity and antibody resistance of the SARS-CoV-2 variant are determined by its mutations on the spike (S) protein receptor-binding domain (RBD). However, a complete experimental evaluation of Omicron might take weeks or even months. Here, we present a comprehensive quantitative analysis of Omicron's infectivity, vaccine-breakthrough, and antibody resistance. An artificial intelligence (AI) model, which has been trained with tens of thousands of experimental data points and extensively validated by experimental data on SARS-CoV-2, reveals that Omicron may be over ten times more contagious than the original virus or about twice as infectious as the Delta variant. Based on 132 three-dimensional (3D) structures of antibody-RBD complexes, we unveil that Omicron may be twice more likely to escape current vaccines than the Delta variant. The Food and Drug Administration (FDA)-approved monoclonal antibodies (mAbs) from Eli Lilly may be seriously compromised. Omicron may also diminish the efficacy of mAbs from Celltrion and Rockefeller University. However, its impact on Regeneron mAb cocktail appears to be mild.
Figures


References
-
- Li Wendong, Shi Zhengli, Yu Meng, Ren Wuze, Smith Craig, Epstein Jonathan H, Wang Hanzhong, Crameri Gary, Hu Zhihong, Zhang Huajun, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science, 310(5748):676–679, 2005. - PubMed
-
- Qu Xiu-Xia, Hao Pei, Song Xi-Jun, Jiang Si-Ming, Liu Yan-Xia, Wang Pei-Gang, Rao Xi, Song Huai-Dong, Wang Sheng-Yue, Zuo Yu, et al. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. Journal of Biological Chemistry, 280(33):29588–29595, 2005. - PMC - PubMed
-
- Song Huai-Dong, Tu Chang-Chun, Zhang Guo-Wei, Wang Sheng-Yue, Zheng Kui, Lei Lian-Cheng, Chen Qiu-Xia, Gao Yu-Wei, Zhou Hui-Qiong, Xiang Hua, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proceedings of the National Academy of Sciences, 102(7):2430–2435, 2005. - PMC - PubMed
-
- Hoffmann Markus, Kleine-Weber Hannah, Schroeder Simon, Krüger Nadine, Herrler Tanja, Erichsen Sandra, Schiergens Tobias S, Herrler Georg, Wu Nai-Huei, Nitsche Andreas, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2):271–280, 2020. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
