Investigating the replicability of preclinical cancer biology
- PMID: 34874005
- PMCID: PMC8651293
- DOI: 10.7554/eLife.71601
Investigating the replicability of preclinical cancer biology
Abstract
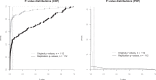
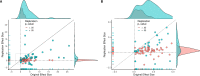
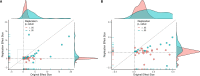
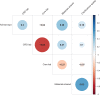
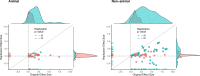
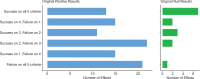
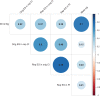
Replicability is an important feature of scientific research, but aspects of contemporary research culture, such as an emphasis on novelty, can make replicability seem less important than it should be. The Reproducibility Project: Cancer Biology was set up to provide evidence about the replicability of preclinical research in cancer biology by repeating selected experiments from high-impact papers. A total of 50 experiments from 23 papers were repeated, generating data about the replicability of a total of 158 effects. Most of the original effects were positive effects (136), with the rest being null effects (22). A majority of the original effect sizes were reported as numerical values (117), with the rest being reported as representative images (41). We employed seven methods to assess replicability, and some of these methods were not suitable for all the effects in our sample. One method compared effect sizes: for positive effects, the median effect size in the replications was 85% smaller than the median effect size in the original experiments, and 92% of replication effect sizes were smaller than the original. The other methods were binary - the replication was either a success or a failure - and five of these methods could be used to assess both positive and null effects when effect sizes were reported as numerical values. For positive effects, 40% of replications (39/97) succeeded according to three or more of these five methods, and for null effects 80% of replications (12/15) were successful on this basis; combining positive and null effects, the success rate was 46% (51/112). A successful replication does not definitively confirm an original finding or its theoretical interpretation. Equally, a failure to replicate does not disconfirm a finding, but it does suggest that additional investigation is needed to establish its reliability.
Keywords: Reproducibility Project: Cancer Biology; cancer biology; computational biology; credibility; human; meta-analysis; mouse; replication; reproducibility; reproducibility in cancer biology; systems biology; transparency.
© 2021, Errington et al.
Conflict of interest statement
TE, AD Employed by the Center for Open Science, a non-profit organization that has a mission to increase openness, integrity, and reproducibility of research, MM No competing interests declared, CS Was employed by the Center for Open Science, a non-profit organization that has a mission to increase openness, integrity, and reproducibility of research, NP Was employed by and holds shares in Science Exchange Inc, EI Employed by and holds shares in Science Exchange Inc, BN Employed by the nonprofit Center for Open Science that has a mission to increase openness, integrity, and reproducibility of research
Figures





Comment in
-
Half of top cancer studies fail high-profile reproducibility effort.Nature. 2021 Dec;600(7889):368-369. doi: 10.1038/d41586-021-03691-0. Nature. 2021. PMID: 34887584 No abstract available.
References
-
- Anderson CJ, Bahník Š, Barnett-Cowan M, Bosco FA, Chandler J, Chartier CR, Cheung F, Christopherson CD, Cordes A, Cremata EJ, Della Penna N, Estel V, Fedor A, Fitneva SA, Frank MC, Grange JA, Hartshorne JK, Hasselman F, Henninger F, van der Hulst M, Jonas KJ, Lai CK, Levitan CA, Miller JK, Moore KS, Meixner JM, Munafò MR, Neijenhuijs KI, Nilsonne G, Nosek BA, Plessow F, Prenoveau JM, Ricker AA, Schmidt K, Spies JR, Stieger S, Strohminger N, Sullivan GB, van Aert RCM, van Assen M, Vanpaemel W, Vianello M, Voracek M, Zuni K. Response to Comment on “Estimating the reproducibility of psychological science.”. Science. 2016;351:1037. doi: 10.1126/science.aad9163. - DOI - PubMed
-
- Andrews I, Kasy M. Identification of and Correction for Publication Bias. American Economic Review. 2019;109:2766–2794. doi: 10.1257/aer.20180310. - DOI
-
- Baker M. Is there a reproducibility crisis. Nature. 2016;533:353–366.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

