Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape
- PMID: 34874007
- PMCID: PMC8651292
- DOI: 10.7554/eLife.73027
Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape
Abstract
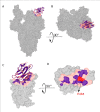
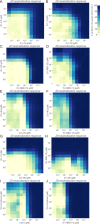
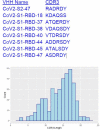
The emergence of SARS-CoV-2 variants threatens current vaccines and therapeutic antibodies and urgently demands powerful new therapeutics that can resist viral escape. We therefore generated a large nanobody repertoire to saturate the distinct and highly conserved available epitope space of SARS-CoV-2 spike, including the S1 receptor binding domain, N-terminal domain, and the S2 subunit, to identify new nanobody binding sites that may reflect novel mechanisms of viral neutralization. Structural mapping and functional assays show that indeed these highly stable monovalent nanobodies potently inhibit SARS-CoV-2 infection, display numerous neutralization mechanisms, are effective against emerging variants of concern, and are resistant to mutational escape. Rational combinations of these nanobodies that bind to distinct sites within and between spike subunits exhibit extraordinary synergy and suggest multiple tailored therapeutic and prophylactic strategies.
Keywords: SARS-CoV-2; human; infectious disease; microbiology; nanobodies; single-domain antibodies; spike glycoprotein; synergy; variants of concern.
© 2021, Mast et al.
Conflict of interest statement
FM, PF, NK, JW, EJ, JO, JA, BC, MR Inventor on a provisional patent describing the anti-spike nanobodies described in this manuscript, TS, KM, FS, MR, YW, LR, EV, ND, VV, SK, JJ, MS, PO, TH, DS, JD, DF, AS, PB No competing interests declared, LH Louis Herlands is affiliated with AbOde Therapeutics Inc. The author has no financial interests to declare
Figures



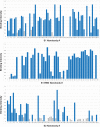

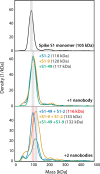

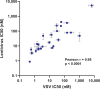
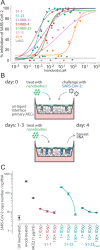




Update of
-
Nanobody Repertoires for Exposing Vulnerabilities of SARS-CoV-2.bioRxiv [Preprint]. 2021 Apr 10:2021.04.08.438911. doi: 10.1101/2021.04.08.438911. bioRxiv. 2021. Update in: Elife. 2021 Dec 07;10:e73027. doi: 10.7554/eLife.73027. PMID: 33851164 Free PMC article. Updated. Preprint.
References
-
- Andreano E, Nicastri E, Paciello I, Pileri P, Manganaro N, Piccini G, Manenti A, Pantano E, Kabanova A, Troisi M, Vacca F, Cardamone D, De Santi C, Torres JL, Ozorowski G, Benincasa L, Jang H, Di Genova C, Depau L, Brunetti J, Agrati C, Capobianchi MR, Castilletti C, Emiliozzi A, Fabbiani M, Montagnani F, Bracci L, Sautto G, Ross TM, Montomoli E, Temperton N, Ward AB, Sala C, Ippolito G, Rappuoli R. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184:1821–1835. doi: 10.1016/j.cell.2021.02.035. - DOI - PMC - PubMed
-
- Bard G. Spelling-Error Tolerant, Order-Independent Pass-Phrases via the Damerau–Levenshtein String-Edit Distance Metric. Darlinghurst, Australia: Australian Computer Society, Inc; 2007.
-
- Barnes CO, Jette CA, Abernathy ME, Dam K-MA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, Robbiani DF, Nussenzweig MC, West AP, Jr, Bjorkman PJ. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

