Nicked Invader probes: multistranded and sequence-unrestricted recognition of double-stranded DNA
- PMID: 34874037
- PMCID: PMC8810728
- DOI: 10.1039/d1ob02019f
Nicked Invader probes: multistranded and sequence-unrestricted recognition of double-stranded DNA
Abstract
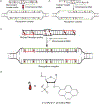
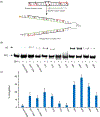
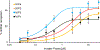
Major efforts have been devoted to the development of constructs that enable sequence-specific recognition of double-stranded (ds) DNA, fueled by the promise for enabling tools for applications in molecular biology, diagnostics, and medicine. Towards this end, we have previously introduced Invader probes, i.e., short DNA duplexes with +1 interstrand zipper arrangements of intercalator-functionalized nucleotides. The individual strands of these labile probes display high affinity towards complementary DNA (cDNA), which drives sequence-unrestricted dsDNA-recognition. However, recognition of long targets is challenging due to the high stability of the corresponding probes. To address this, we recently introduced toehold Invader probes, i.e., Invader probes with 5'-single-stranded overhangs. The toehold architecture allows for shorter double-stranded segments to be used, which facilitates probe dissociation and dsDNA-recognition. As an extension thereof, we here report the biophysical and dsDNA-targeting properties of nicked Invader probes. In this probe architecture, the single-stranded overhangs of toehold Invader probes are hybridized to short intercalator-modified auxiliary strands, leading to formation of additional labile segments. The extra binding potential from the auxiliary strands imparts nicked Invader probes with greater dsDNA-affinity than the corresponding toehold or blunt-ended probes. Recognition of chromosomal DNA targets, refractory to recognition by conventional Invader probes, is demonstrated for nicked Invader probes in the context of non-denaturing FISH experiments, which highlights their utility as dsDNA-targeting tools.
Conflict of interest statement
CONFLICTS OF INTERESTS
P. J. H. is an inventor on patents pertaining to Invader probes, which have been issued to the University Idaho.
Figures


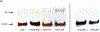
Similar articles
-
Recognition of double-stranded DNA using LNA-modified toehold Invader probes.Org Biomol Chem. 2021 Nov 3;19(42):9276-9290. doi: 10.1039/d1ob01888d. Org Biomol Chem. 2021. PMID: 34657934 Free PMC article.
-
Mixed-Sequence Recognition of Double-Stranded DNA Using Enzymatically Stable Phosphorothioate Invader Probes.Molecules. 2015 Jul 29;20(8):13780-93. doi: 10.3390/molecules200813780. Molecules. 2015. PMID: 26230684 Free PMC article.
-
Impact of non-nucleotidic bulges on recognition of mixed-sequence dsDNA by pyrene-functionalized Invader probes.Org Biomol Chem. 2020 Jun 24;18(24):4645-4655. doi: 10.1039/d0ob01052a. Org Biomol Chem. 2020. PMID: 32520054 Free PMC article.
-
DNA probes: applications of the principles of nucleic acid hybridization.Crit Rev Biochem Mol Biol. 1991;26(3-4):227-59. doi: 10.3109/10409239109114069. Crit Rev Biochem Mol Biol. 1991. PMID: 1718662 Review.
-
Revolving hexameric ATPases as asymmetric motors to translocate double-stranded DNA genome along one strand.iScience. 2023 May 19;26(6):106922. doi: 10.1016/j.isci.2023.106922. eCollection 2023 Jun 16. iScience. 2023. PMID: 37305704 Free PMC article. Review.
References
-
- Hari Y, Obika S and Imanishi T, Eur. J. Org. Chem, 2012, 2012, 2875–2887.
-
- Kaihatsu K, Janowski BA and Corey DR, Chem. Biol, 2004, 11, 749–758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

