No one tool to rule them all: prokaryotic gene prediction tool annotations are highly dependent on the organism of study
- PMID: 34875010
- PMCID: PMC8825762
- DOI: 10.1093/bioinformatics/btab827
No one tool to rule them all: prokaryotic gene prediction tool annotations are highly dependent on the organism of study
Abstract
Motivation: The biases in CoDing Sequence (CDS) prediction tools, which have been based on historic genomic annotations from model organisms, impact our understanding of novel genomes and metagenomes. This hinders the discovery of new genomic information as it results in predictions being biased towards existing knowledge. To date, users have lacked a systematic and replicable approach to identify the strengths and weaknesses of any CDS prediction tool and allow them to choose the right tool for their analysis.
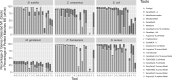
Results: We present an evaluation framework (ORForise) based on a comprehensive set of 12 primary and 60 secondary metrics that facilitate the assessment of the performance of CDS prediction tools. This makes it possible to identify which performs better for specific use-cases. We use this to assess 15 ab initio- and model-based tools representing those most widely used (historically and currently) to generate the knowledge in genomic databases. We find that the performance of any tool is dependent on the genome being analysed, and no individual tool ranked as the most accurate across all genomes or metrics analysed. Even the top-ranked tools produced conflicting gene collections, which could not be resolved by aggregation. The ORForise evaluation framework provides users with a replicable, data-led approach to make informed tool choices for novel genome annotations and for refining historical annotations.
Availability and implementation: Code and datasets for reproduction and customisation are available at https://github.com/NickJD/ORForise.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press.
Figures

References
-
- Al-Turaiki, Israa M., et al. "Computational approaches for gene prediction: a comparative survey." International Conference on Informatics Engineering and Information Science. Springer, Berlin, Heidelberg, 2011.
-
- Andrews S.J., Rothnagel J.A. (2014) Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet., 15, 193–204. - PubMed
-
- Badger J.H., Olsen G.J. (1999) CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol., 16, 512–524. - PubMed
-
- Baranov P.V. et al. (2015) Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nat. Rev. Genet., 16, 517–529. - PubMed
Publication types
MeSH terms
Grants and funding
- Institute of Biological, Environmental and Rural Sciences Aberystwyth PhD fellowship
- BB/E/W/10964A01/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- Department of Agriculture, Food and the Marine Ireland/DAERA Northern Ireland
- European Commission via Horizon 2020
- BBS/OS/GC/000011B/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom

