Biological heterogeneity in idiopathic pulmonary arterial hypertension identified through unsupervised transcriptomic profiling of whole blood
- PMID: 34876579
- PMCID: PMC8651638
- DOI: 10.1038/s41467-021-27326-0
Biological heterogeneity in idiopathic pulmonary arterial hypertension identified through unsupervised transcriptomic profiling of whole blood
Erratum in
-
Author Correction: Biological heterogeneity in idiopathic pulmonary arterial hypertension identified through unsupervised transcriptomic profiling of whole blood.Nat Commun. 2022 Nov 25;13(1):7276. doi: 10.1038/s41467-022-33381-y. Nat Commun. 2022. PMID: 36433938 Free PMC article. No abstract available.
Abstract
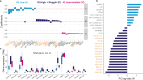
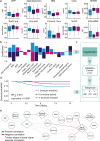
Idiopathic pulmonary arterial hypertension (IPAH) is a rare but fatal disease diagnosed by right heart catheterisation and the exclusion of other forms of pulmonary arterial hypertension, producing a heterogeneous population with varied treatment response. Here we show unsupervised machine learning identification of three major patient subgroups that account for 92% of the cohort, each with unique whole blood transcriptomic and clinical feature signatures. These subgroups are associated with poor, moderate, and good prognosis. The poor prognosis subgroup is associated with upregulation of the ALAS2 and downregulation of several immunoglobulin genes, while the good prognosis subgroup is defined by upregulation of the bone morphogenetic protein signalling regulator NOG, and the C/C variant of HLA-DPA1/DPB1 (independently associated with survival). These findings independently validated provide evidence for the existence of 3 major subgroups (endophenotypes) within the IPAH classification, could improve risk stratification and provide molecular insights into the pathogenesis of IPAH.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 205188/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- FS/18/52/33808/BHF_/British Heart Foundation/United Kingdom
- DH_/Department of Health/United Kingdom
- SP/12/12/29836/BHF_/British Heart Foundation/United Kingdom
- SP/14/6/31350/BHF_/British Heart Foundation/United Kingdom
- PG/11/116/29288/BHF_/British Heart Foundation/United Kingdom
- CH/09/001/25945/BHF_/British Heart Foundation/United Kingdom
- RE/18/4/34215/BHF_/British Heart Foundation/United Kingdom
- FS/18/13/33281/BHF_/British Heart Foundation/United Kingdom
- MR/M008894/1/MRC_/Medical Research Council/United Kingdom
- R01 HL136603/HL/NHLBI NIH HHS/United States
- MR/K020919/1/MRC_/Medical Research Council/United Kingdom
- FS/15/59/31839/BHF_/British Heart Foundation/United Kingdom
- SP/18/10/33975/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

