Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2
- PMID: 34876592
- PMCID: PMC8651658
- DOI: 10.1038/s41467-021-27318-0
Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2
Abstract
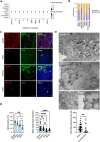
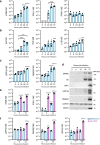
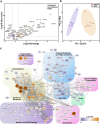
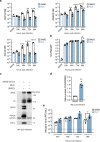
The nasal epithelium is a plausible entry point for SARS-CoV-2, a site of pathogenesis and transmission, and may initiate the host response to SARS-CoV-2. Antiviral interferon (IFN) responses are critical to outcome of SARS-CoV-2. Yet little is known about the interaction between SARS-CoV-2 and innate immunity in this tissue. Here we apply single-cell RNA sequencing and proteomics to a primary cell model of human nasal epithelium differentiated at air-liquid interface. SARS-CoV-2 demonstrates widespread tropism for nasal epithelial cell types. The host response is dominated by type I and III IFNs and interferon-stimulated gene products. This response is notably delayed in onset relative to viral gene expression and compared to other respiratory viruses. Nevertheless, once established, the paracrine IFN response begins to impact on SARS-CoV-2 replication. When provided prior to infection, recombinant IFNβ or IFNλ1 induces an efficient antiviral state that potently restricts SARS-CoV-2 viral replication, preserving epithelial barrier integrity. These data imply that the IFN-I/III response to SARS-CoV-2 initiates in the nasal airway and suggest nasal delivery of recombinant IFNs to be a potential chemoprophylactic strategy.
© 2021. The Author(s).
Conflict of interest statement
S.H. declares honoraria from CSL Behring and Takeda for teaching and consultancy. M.B. declares being CI on investigator-led research grants from Pfizer and Roche Diagnostics; speaker fees paid to Newcastle University from Novartis, Roche Diagnostics and TEVA; travel expenses to educational meetings from Boehringer Ingelheim and Vertex. The remaining authors declare no competing interests.
Figures


References
-
- Group, R. C. et al. Dexamethasone in hospitalized patients with Covid-19—preliminary report. N. Engl. J. Med. 10.1056/NEJMoa2021436 (2020).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

