A computational pipeline for functional gene discovery
- PMID: 34876638
- PMCID: PMC8651667
- DOI: 10.1038/s41598-021-03041-0
A computational pipeline for functional gene discovery
Abstract
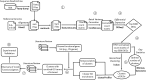
Many computational pipelines exist for the detection of differentially expressed genes. However, computational pipelines for functional gene detection rarely exist. We developed a new computational pipeline for functional gene identification from transcriptome profiling data. Key features of the pipeline include batch effect correction, clustering optimization by gap statistics, gene ontology analysis of clustered genes, and literature analysis for functional gene discovery. By leveraging this pipeline on RNA-seq datasets from two mouse retinal development studies, we identified 7 candidate genes involved in the formation of the photoreceptor outer segment. The expression of top three candidate genes (Pde8b, Laptm4b, and Nr1h4) in the outer segment of the developing mouse retina were experimentally validated by immunohistochemical analysis. This computational pipeline can accurately predict novel functional gene for a specific biological process, e.g., development of the outer segment and synapses of the photoreceptor cells in the mouse retina. This pipeline can also be useful to discover functional genes for other biological processes and in other organs and tissues.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

