Exploring Prognosis-Associated Biomarkers of Estrogen-Independent Uterine Corpus Endometrial Carcinoma by Bioinformatics Analysis
- PMID: 34876842
- PMCID: PMC8643178
- DOI: 10.2147/IJGM.S341345
Exploring Prognosis-Associated Biomarkers of Estrogen-Independent Uterine Corpus Endometrial Carcinoma by Bioinformatics Analysis
Abstract
Background: Uterine corpus endometrial carcinoma (UCEC) is one of the most common female cancers with high incidence and mortality rates. In particular, the prognosis of type II UCEC is poorer than that of type I. However, the molecular mechanism underlying type II UCEC remains unclear.
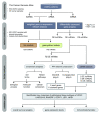
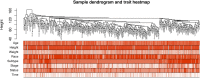
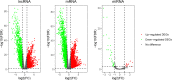
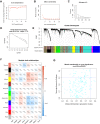
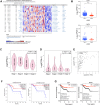
Methods: RNA-seq data and corresponding clinical information on UCEC patients were downloaded from The Cancer Genome Atlas database, which were then separated into mRNA, lncRNA, and miRNA gene expression profile matrix to perform differentially expressed gene analysis. Weighted gene co-expression network analysis (WGCNA) was used to identify key modules associated with different UCEC subtypes based on mRNA and lncRNA expression matrix. Following that, a subtype-associated competing endogenous RNA (ceRNA) regulatory network was constructed. In addition, GO functional annotation and KEGG pathway analysis were performed on subtype-related DE mRNAs, and STRING database was utilized to predict the interaction network between proteins and their biological functions. The key mRNAs were validated at the protein and gene expression levels in endometrial cancerous tissues as compared with normal tissues.
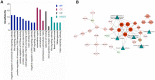
Results: In summary, we identified 4611 mRNA, 3568 lncRNAs, and 47 miRNAs as differentially expressed between endometrial cancerous tissues and normal endometrial tissues. WGCNA demonstrated that 72 mRNAs and 55 lncRNAs were correlated with pathological subtypes. In the constructed ceRNA regulatory network, LINC02418, RASGRF1, and GCNT1 were screened for their association with poor prognosis of type II UCEC. These DE mRNAs were linked to Wnt signaling pathway, and lower expression of LEF1 and NKD1 predicted advanced clinical stages and worse prognosis of UCEC patients.
Conclusion: This study revealed five prognosis-associated biomarkers that can be used to predict the worst prognosis of type II UCEC.
Keywords: WGCNA; competing endogenous RNA; estrogen-independent; prognosis; uterine corpus endometrial carcinoma.
© 2021 Ye et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

