Tracking the amino acid changes of spike proteins across diverse host species of severe acute respiratory syndrome coronavirus 2
- PMID: 34877480
- PMCID: PMC8638202
- DOI: 10.1016/j.isci.2021.103560
Tracking the amino acid changes of spike proteins across diverse host species of severe acute respiratory syndrome coronavirus 2
Abstract
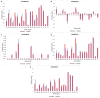
Knowledge of the host-specific properties of the spike protein is of crucial importance to understand the adaptability of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) to infect multiple species and alter transmissibility, particularly in humans. Here, we propose a spike protein predictor SPIKES incorporating with an inheritable bi-objective combinatorial genetic algorithm to identify the biochemical properties of spike proteins and determine their specificity to human hosts. SPIKES identified 20 informative physicochemical properties of the spike protein, including information measures for alpha helix and relative mutability, and amino acid and dipeptide compositions, which have shown compositional difference at the amino acid sequence level between human and diverse animal coronaviruses. We suggest that alterations of these amino acids between human and animal coronaviruses may provide insights into the development and transmission of SARS-CoV-2 in human and other species and support the discovery of targeted antiviral therapies.
Keywords: Association analysis; Bioinformatics; Virology.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
SPIKES: Identification of physicochemical properties of spike proteins across diverse host species of SARS-CoV-2.STAR Protoc. 2022 May 24;3(3):101460. doi: 10.1016/j.xpro.2022.101460. eCollection 2022 Sep 16. STAR Protoc. 2022. PMID: 35726315 Free PMC article.
-
Cryo-electron Microscopy Structure of the Swine Acute Diarrhea Syndrome Coronavirus Spike Glycoprotein Provides Insights into Evolution of Unique Coronavirus Spike Proteins.J Virol. 2020 Oct 27;94(22):e01301-20. doi: 10.1128/JVI.01301-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32817223 Free PMC article.
-
Dynamics of SARS-CoV-2 Spike Proteins in Cell Entry: Control Elements in the Amino-Terminal Domains.mBio. 2021 Aug 31;12(4):e0159021. doi: 10.1128/mBio.01590-21. Epub 2021 Aug 3. mBio. 2021. PMID: 34340537 Free PMC article.
-
A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses.Emerg Microbes Infect. 2020 Mar 14;9(1):558-570. doi: 10.1080/22221751.2020.1736644. eCollection 2020. Emerg Microbes Infect. 2020. PMID: 32172672 Free PMC article. Review.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
Cited by
-
Predicting host species susceptibility to influenza viruses and coronaviruses using genome data and machine learning: a scoping review.Front Vet Sci. 2024 Sep 25;11:1358028. doi: 10.3389/fvets.2024.1358028. eCollection 2024. Front Vet Sci. 2024. PMID: 39386249 Free PMC article.
-
Sequence analysis of the Spike, RNA-dependent RNA polymerase, and protease genes reveals a distinct evolutionary pattern of SARS-CoV-2 variants circulating in Yogyakarta and Central Java provinces, Indonesia.Virus Genes. 2024 Apr;60(2):105-116. doi: 10.1007/s11262-023-02048-1. Epub 2024 Jan 20. Virus Genes. 2024. PMID: 38244104
-
Computational and comparative investigation of hydrophobic profile of spike protein of SARS-CoV-2 and SARS-CoV.J Biol Phys. 2022 Dec;48(4):399-414. doi: 10.1007/s10867-022-09615-x. Epub 2022 Nov 23. J Biol Phys. 2022. PMID: 36422744 Free PMC article.
-
A review on evolution of emerging SARS-CoV-2 variants based on spike glycoprotein.Int Immunopharmacol. 2022 Apr;105:108565. doi: 10.1016/j.intimp.2022.108565. Epub 2022 Jan 29. Int Immunopharmacol. 2022. PMID: 35123183 Free PMC article. Review.
-
Decoy peptides effectively inhibit the binding of SARS-CoV-2 to ACE2 on oral epithelial cells.Heliyon. 2023 Nov 20;9(12):e22614. doi: 10.1016/j.heliyon.2023.e22614. eCollection 2023 Dec. Heliyon. 2023. PMID: 38107325 Free PMC article.
References
-
- Arora N., Banerjee A.K., Narasu M.L. The role of artificial intelligence in tackling COVID-19. Future Virol. 2020 doi: 10.2217/fvl-2020-0130. - DOI
-
- Benjamini Y., Krieger A.M., Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93:491–507.
LinkOut - more resources
Full Text Sources
Miscellaneous

