Proteome and strain analysis of cyanobacterium Candidatus "Phormidium alkaliphilum" reveals traits for success in biotechnology
- PMID: 34877483
- PMCID: PMC8633866
- DOI: 10.1016/j.isci.2021.103405
Proteome and strain analysis of cyanobacterium Candidatus "Phormidium alkaliphilum" reveals traits for success in biotechnology
Abstract
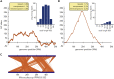
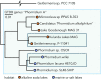
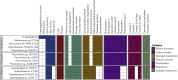
Cyanobacteria encompass a diverse group of photoautotrophic bacteria with important roles in nature and biotechnology. Here we characterized Candidatus "Phormidium alkaliphilum," an abundant member in alkaline soda lake microbial communities globally. The complete, circular whole-genome sequence of Ca. "P. alkaliphilum" was obtained using combined Nanopore and Illumina sequencing of a Ca. "P. alkaliphilum" consortium. Strain-level diversity of Ca. "P. alkaliphilum" was shown to contribute to photobioreactor robustness under different operational conditions. Comparative genomics of closely related species showed that adaptation to high pH was not attributed to specific genes. Proteomics at high and low pH showed only minimal changes in gene expression, but higher productivity in high pH. Diverse photosystem antennae proteins, and high-affinity terminal oxidase, compared with other soda lake cyanobacteria, appear to contribute to the success of Ca. "P. alkaliphilum" in photobioreactors and biotechnology applications.
Keywords: Cyanobacteria; Genomics; Microbial biotechnology; Proteomics.
© 2021.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Almon H., Böger P. Hydrogen metabolism of the unicellular cyanobacterium Chroococcidiopsis thermalis ATCC29380. FEMS Microbiol. Lett. 1988;49:445–449.
-
- Badger M.R., Price G.D. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 2003;54:609–622. - PubMed
LinkOut - more resources
Full Text Sources

