STING regulates peripheral nerve regeneration and colony stimulating factor 1 receptor (CSF1R) processing in microglia
- PMID: 34877494
- PMCID: PMC8633968
- DOI: 10.1016/j.isci.2021.103434
STING regulates peripheral nerve regeneration and colony stimulating factor 1 receptor (CSF1R) processing in microglia
Abstract
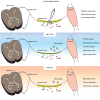
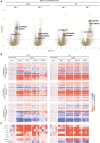
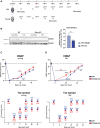
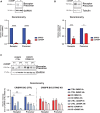
Inflammatory responses are crucial for regeneration following peripheral nerve injury (PNI). PNI triggers inflammatory responses at the site of injury. The DNA-sensing receptor cyclic GMP-AMP synthase (cGAS) and its downstream effector stimulator of interferon genes (STING) sense foreign and self-DNA and trigger type I interferon (IFN) immune responses. We demonstrate here that following PNI, the cGAS/STING pathway is upregulated in the sciatic nerve of naive rats and dysregulated in old rats. In a nerve crush mouse model where STING is knocked out, myelin content in sciatic nerve is increased resulting in accelerated functional axon recovery. STING KO mice have lower macrophage number in sciatic nerve and decreased microglia activation in spinal cord 1 week post injury. STING activation regulated processing of colony stimulating factor 1 receptor (CSF1R) and microglia survival in vitro. Taking together, these data highlight a previously unrecognized role of STING in the regulation of nerve regeneration.
Keywords: Cell biology; Immunology; Molecular biology; Neuroscience.
© 2021 The Authors.
Conflict of interest statement
All authors are employees and some are shareholders of Novartis.
Figures



References
-
- Baccarini M., Dello Sbarba P., Buscher D., Bartocci A., Stanley E.R. IFN-gamma/lipopolysaccharide activation of macrophages is associated with protein kinase C-dependent down-modulation of the colony-stimulating factor-1 receptor. J. Immunol. 1992;149:2656–2661. - PubMed
-
- Barrette B., Calvo E., Vallieres N., Lacroix S. Transcriptional profiling of the injured sciatic nerve of mice carrying the Wld(S) mutant gene: Identification of genes involved in neuroprotection, neuroinflammation, and nerve regeneration. Brain Behav. Immun. 2010;24:1254–1267. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

