Retinoic acid signaling drives differentiation toward the absorptive lineage in colorectal cancer
- PMID: 34877501
- PMCID: PMC8633980
- DOI: 10.1016/j.isci.2021.103444
Retinoic acid signaling drives differentiation toward the absorptive lineage in colorectal cancer
Abstract
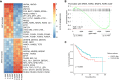
Retinoic acid (RA) signaling is an important and conserved pathway that regulates cellular proliferation and differentiation. Furthermore, perturbed RA signaling is implicated in cancer initiation and progression. However, the mechanisms by which RA signaling contributes to homeostasis, malignant transformation, and disease progression in the intestine remain incompletely understood. Here, we report, in agreement with previous findings, that activation of the Retinoic Acid Receptor and the Retinoid X Receptor results in enhanced transcription of enterocyte-specific genes in mouse small intestinal organoids. Conversely, inhibition of this pathway results in reduced expression of genes associated with the absorptive lineage. Strikingly, this latter effect is conserved in a human organoid model for colorectal cancer (CRC) progression. We further show that RXR motif accessibility depends on progression state of CRC organoids. Finally, we show that reduced RXR target gene expression correlates with worse CRC prognosis, implying RA signaling as a putative therapeutic target in CRC.
Keywords: Biological sciences; Cancer; Cell biology; Functional aspects of cell biology; Oncology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Agarwal C., Chandraratna R.A.S., Johnson A.T., Rorke E.A., Eckert R.L. AGN193109 is a highly effective antagonist of retinoid action in human ectocervical epithelial cells (∗) J. Biol. Chem. 1996;271:12209–12212. - PubMed
-
- Auclair B.A., Benoit Y.D., Rivard N., Mishina Y., Perreault N. Bone morphogenetic protein signaling is essential for terminal differentiation of the intestinal secretory cell lineage. Gastroenterology. 2007;133:887–896. - PubMed
-
- Bruse N., van Heeringen S.J. GimmeMotifs: an analysis framework for transcription factor motif analysis. bioRxiv. 2018:474403.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

