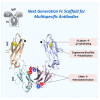
Next generation Fc scaffold for multispecific antibodies
- PMID: 34877503
- PMCID: PMC8633962
- DOI: 10.1016/j.isci.2021.103447
Next generation Fc scaffold for multispecific antibodies
Abstract
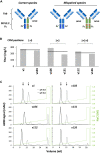
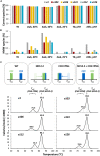
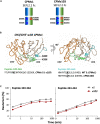
Bispecific antibodies (Bispecifics) demonstrate exceptional clinical potential to address some of the most complex diseases. However, Bispecific production in a single cell often requires the correct pairing of multiple polypeptide chains for desired assembly. This is a considerable hurdle that hinders the development of many immunoglobulin G (IgG)-like bispecific formats. Our approach focuses on the rational engineering of charged residues to facilitate the chain pairing of distinct heavy chains (HC). Here, we deploy structure-guided protein design to engineer charge pair mutations (CPMs) placed in the CH3-CH3' interface of the fragment crystallizable (Fc) region of an antibody (Ab) to correctly steer heavy chain pairing. When used in combination with our stable effector functionless 2 (SEFL2.2) technology, we observed high pairing efficiency without significant losses in expression yields. Furthermore, we investigate the relationship between CPMs and the sequence diversity in the parental antibodies, proposing a rational strategy to deploy these engineering technologies.
Keywords: Biochemistry; Bioengineering; Biomolecular engineering; Structural biology.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Atwell S., Ridgway J.B., Wells J.A., Carter P. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J. Mol. Biol. 1997;270:26–35. - PubMed
-
- Backliwal G., Hildinger M., Kuettel I., Delegrange F., Hacker D.L., Wurm F.M. Valproic acid: A viable alternative to sodium butyrate for enhancing protein expression in mammalian cell cultures. Biotechnol. Bioeng. 2008;101:182–189. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

